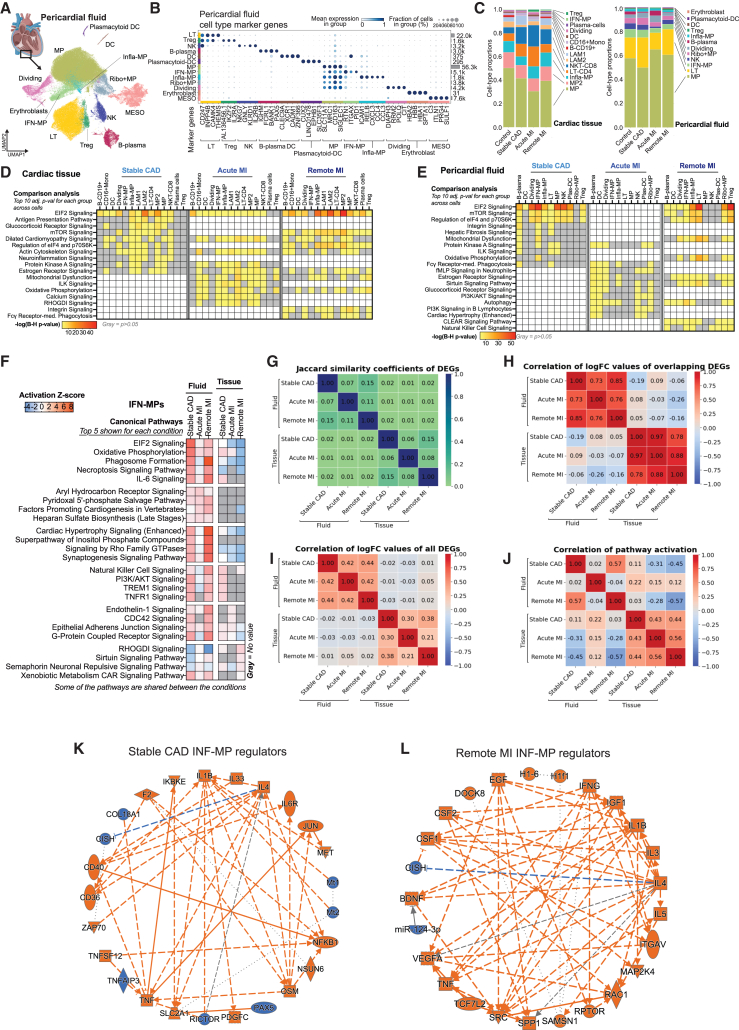
Figure 5.
Pericardial fluid cells reflect the changes in disease states
(A and B) (A) snRNA-seq data and (B) marker genes for immune cell subtypes in pericardial fluid. Heart image by BioRender.com.
(C) Immune cell proportions in cardiac tissue and corresponding pericardial fluid samples.
(D and E) Top 10 pathways with highest significance score in IPA comparison analysis (B–H adjusted p value <0.05) in cardiac tissue (D) and corresponding pericardial fluid samples (E) in stable CAD (N = 5), acute MI (N = 4), and remote MI (N = 5) compared to control (N = 4). Only the top 10 for each group are shown. Full data can be explored at http://compbio2.mit.edu/scheart/.
(F) Top pathways based on activation Z score from IPA for each condition in fluid and tissue IFN-MPs. Positive values indicate pathway activation based on underlying gene expression patterns and blue pathway inhibition.
(G) Jaccard similarity coefficiency for differentially expressed genes (DEGs) in IFN-MPs.
(H) Correlation of log fold change (logFC) values of overlapping DEGs (i.e., found in all compared groups) in IFN-MPs.
(I) Correlation of logFC values of all DEGs in IFN-MPs.
(J) Correlation of activation Z scores for enriched pathways in IFN-MPs.
(K and L) Regulator networks for stable CAD and remote MI in IFN-MPs from IPA’s graphical summary.