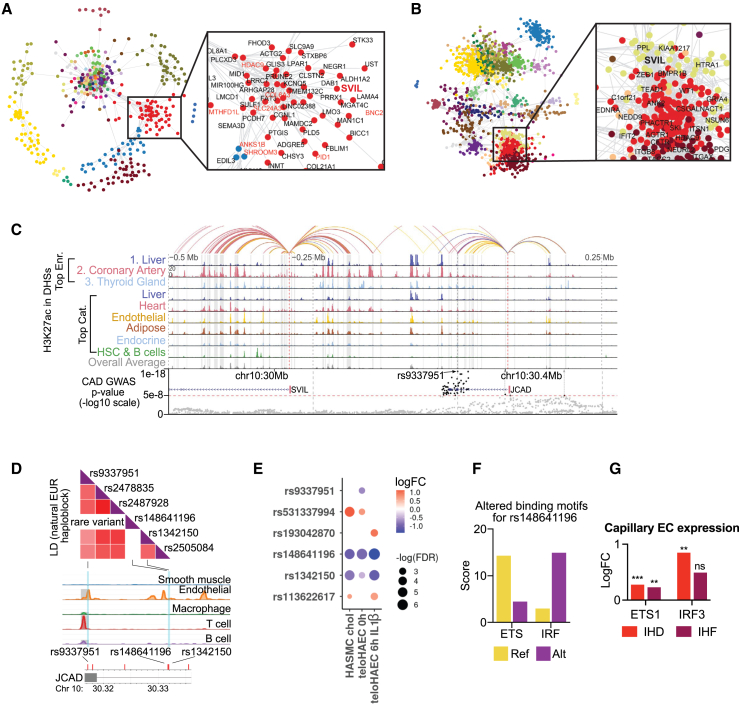
Figure 7.
Dissection of JCAD/SVIL locus
(A and B) Gene expression module for SVIL in SMCs (A) and pericardial fluid cells (B). GWAS-linked genes in red.
(C) Epimap linking of the JCAD/SVIL locus.
(D) Illustration of LD SNPs of the natural European haploblock combined with the single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) data from human coronary arteries generated using LDlink.
(E) Allele-specific enhancer activity measured with STARR-seq in teloHAECs under basal and inflammatory conditions (6 h IL-1β) and HASMCs subjected to cholesterol loading for 24 h. SNPs demonstrating significant changes (false discovery rate [FDR] <0.1) in enhancer activity.
(F) Transcription factor binding motifs altered by rs148641196. Position weight matrix scores shown for reference and alternate.
(G) Changes in transcription factor ETS1 and IRF3 gene expression in capillary ECs in IHD and IHF in snRNA-seq by Nebula. (∗∗p < 0.01, ∗∗∗p < 0.001)