Figure 3.
Cryo-EM structure of EBV gH/gL/gp42-2C1 Fab and mutation verification
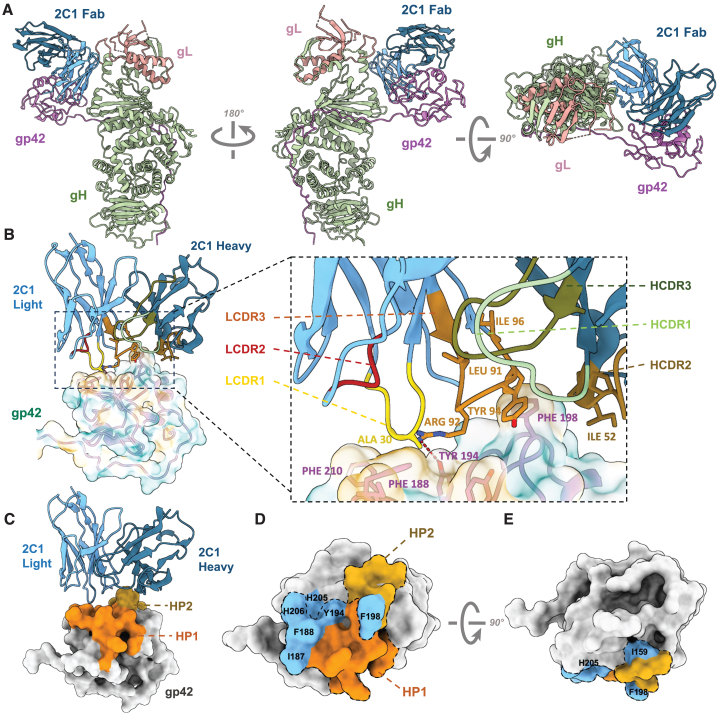
(A) Structure of EBV gH/gL/gp42-2C1 Fab complex. gH, gL, gp42, 2C1 Fab heavy chain, and 2C1 Fab light chain are colored green, salmon, purple, dark blue, and light blue, respectively.
(B) Structure model and zoomed-in views of the binding site of 2C1 Fab (ribbon representation). The CDRs of the vH (HCDR1-3) and the CDRs of the vL (LCDR1-3) are shown in different colors. The side chains of residues that form the hydrophobic surface (yellow: hydrophobic amino acids, blue: hydrophilic amino acids) of gp42 are displayed.
(C) Structure model of 2C1 Fab (ribbon representation) with EBV gp42 (surface representation). Hydrophobic patches (HPs) are indicated.
(D and E) Different views of overlap between primary functional amino acids and HPs. Residues Ile159, Ile187, Phe188, Tyr194, Phe198, His205, and His206 of the HPs are indicated in blue.
See also Figures S3–S5 and Table S1.