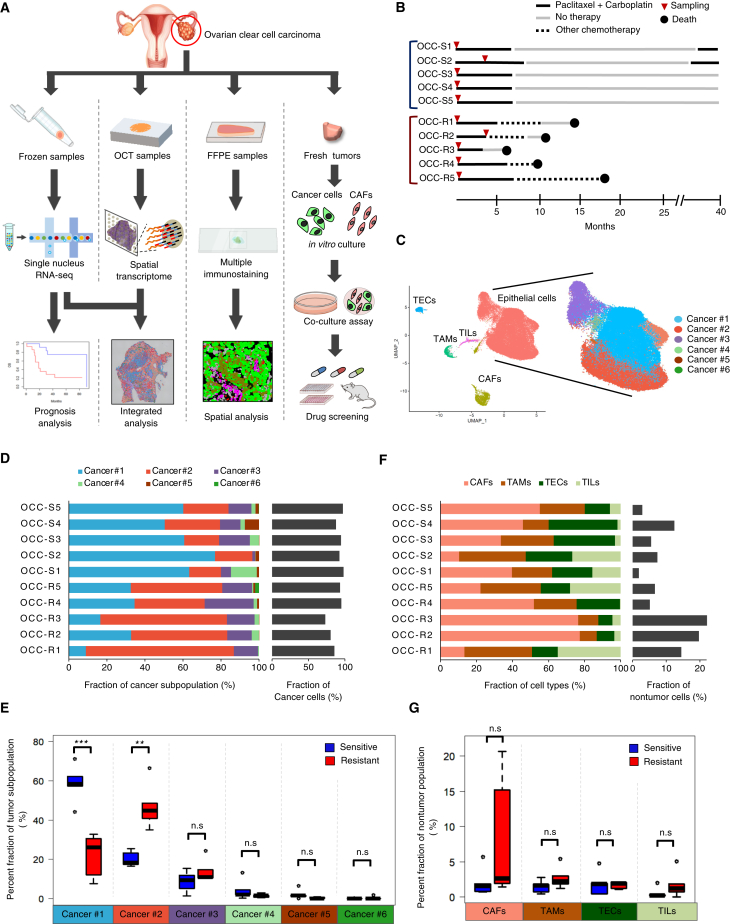
Figure 1.
Identification of a cancer cell subpopulation associated with chemoresistance of OCCC
(A) Study design based on surgical specimens of OCCC.
(B) Timeline presentation of 10 patients whose tumors were used for snRNA-seq, spatial transcriptome, and immunostaining analyses.
(C) UMAP presentation of snRNA-seq data obtained from chemoresistant (OCC-R1-5) and chemosensitive (OCC-S1-5) OCCC. Original snRNA-seq data from the 10 tumors (Figure S1A) were subjected to anchoring prior to data integration. Subsequently, the identity of cells in each cluster was determined based upon expression of marker genes,17 as shown in Figure S1C. TEC, tumor endothelial cells; CAF, cancer-associated fibroblast; TAM, tumor-associated macrophage; TIL, tumor-infiltrating lympohcyte. The epithelial tumor population was further classified into six subpopulations (Cancer #1–6), denoted by the indicated colors.
(D) Stacked bar graph displaying the distribution of each cancer cell subpopulation (Cancer #1–6) in each tumor (left) and the percentage of cancer cells (right).
(E) Boxplots showing the fraction of the indicated cancer subpopulations in the chemoresistant and chemosensitive tumors shown in (D). The horizontal bars within the boxes indicate the median value. The top and bottom bars of the box denote the 25th and 75th percentiles, respectively. ∗∗∗p < 0.001; n.s., not significant.
(F) Stacked bar graph displaying the distribution of non-tumor cell populations within each tumor (left) and the percentage of non-tumor cells (right).
(G) Boxplots showing the fractions of the indicated non-tumor populations. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. p values were determined by Student’s t test.