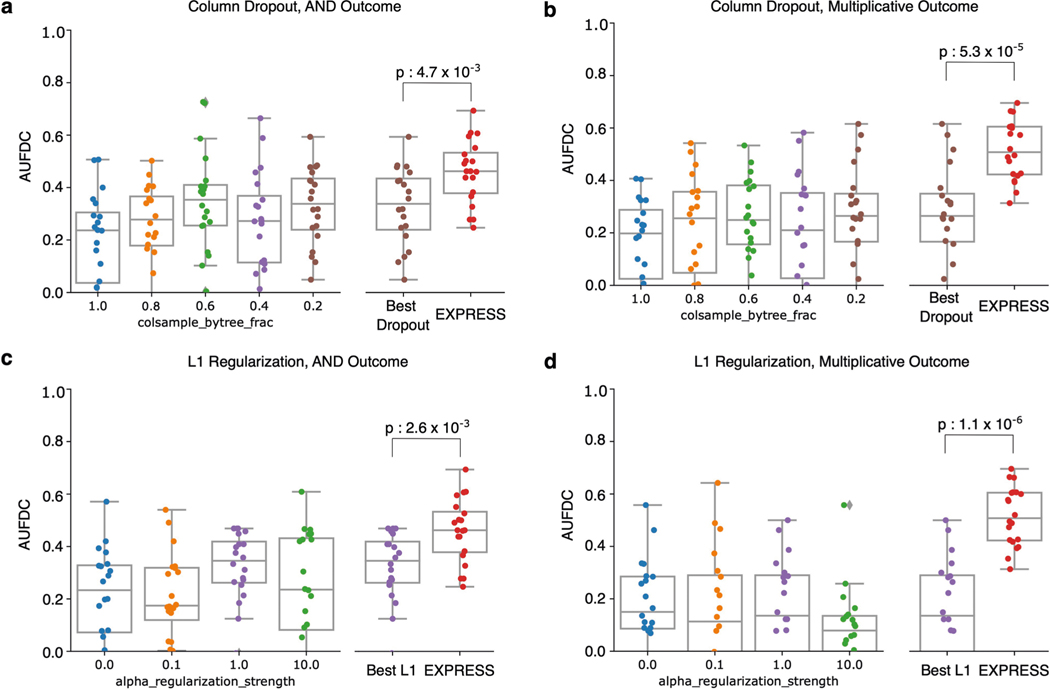
Extended Data Fig. 10 |. Ensembling improves XGBoost attributions more than explicit regularization.
Using the synthetic datasets with real AML gene expression features, we compare the increase in AUFDC seen with explicit regularization, such as per-tree column dropout and L1 regularization, with ensembling. For the synthetic datasets with AML features and the AND true function, we see that ensembles improve AUFDC significantly more than column dropout (a, two-sided Mann−Whitney -test, , ) and L1 regularization (c, , ). For the synthetic datasets with AML features and the multiplicative true function, we see that ensembles improve AUFDC significantly more than column dropout (b, , 5.25 × 10−5) and L1 regularization (d, , ).