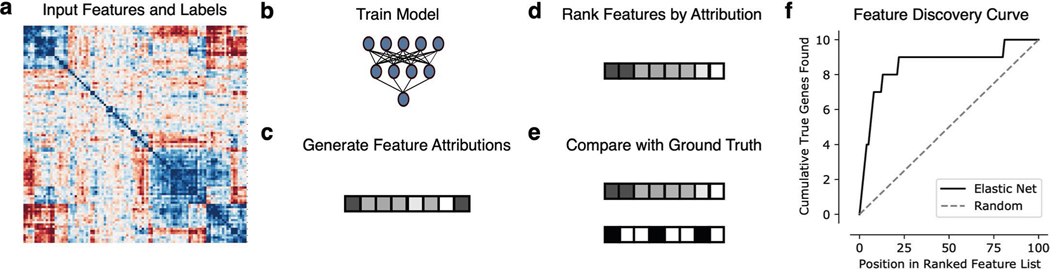
Extended Data Fig. 2 |. Feature discovery benchmark.
For each synthetic or semi-synthetic dataset (a), we trained a variety of models (b) including neural networks, GBMs, support vector machines, and elastic net regression, as well as univariate statistics (Pearson correlation). For the machine-learning models, we then used SAGE to generate global Shapley value feature attributions (c), ranked the features according to the magnitude of their attributions (d), and compared the ranked list generated by each method to the binary ground truth importance vector (e). To measure the feature discovery quality of each method, we plotted how many “true” features are found cumulatively at each point in the ranked feature list (f), then summarized the curve generated by this procedure by measuring the AUFDC. This score is then rescaled so that a score of 0 represents random performance while a score of 1 represents perfect performance.