Fig. 7: DUXBL and TRIM24/TRIM33 co-localize with H3K9me3 deposition.
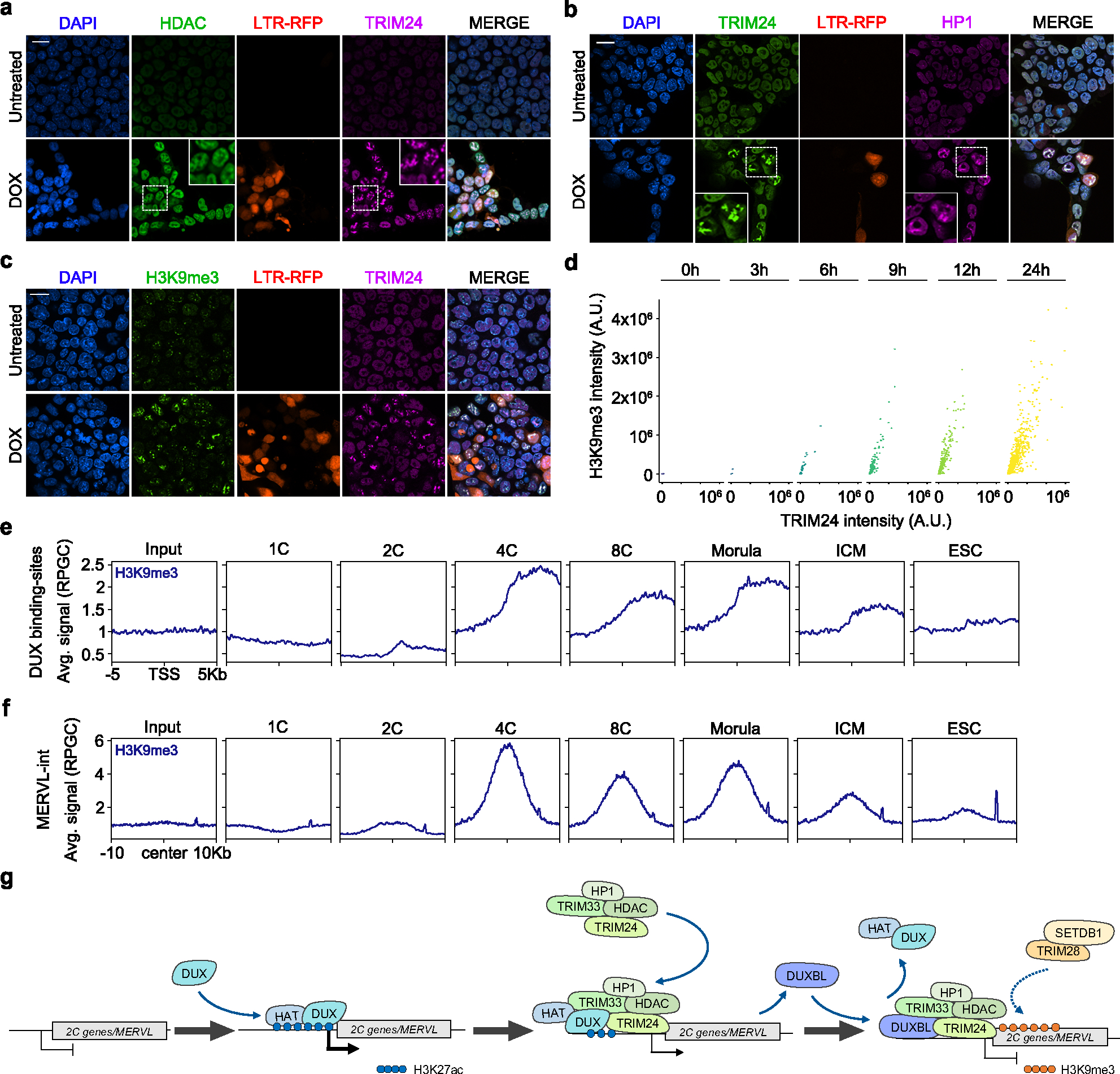
a, b) Immunofluorescence analysis of HDAC (a) and HP1 (b) in untreated or DOX-treated LTR-RFP reporter ESCDUX. DAPI was used to visualize nuclei. Scale bars, 20 μm. Dashed regions are magnified in the insets. c) Immunofluorescence analysis of H3K9me3 in untreated or DOX-treated LTR-RFP reporter ESCDUX. DAPI was used to visualize nuclei. Scale bars, 20 μm. d) Graphs showing TRIM24 and H3K9me3 fluorescence intensity at TRIM24-foci at different times after the addition of DOX in ESCDUX. e, f) ChIPseq read density plot (RPGC) showing H3K9me3 enrichment at DUX-bound sites (e) and MERVL (f) elements occupied by DUXBL after DUX expression during embryonic development. Input (IgG) is shown as reference control. ChIP-seq data obtained from40. g) Schematic representation of the model showing the role of DUXBL facilitating the silencing of DUX-induced transcription. TRIM24/TRIM33 interact with TRIM28 which by recruiting SETDB1 could mediate H3K9me3 deposition. At least two independent experiments using two ESCDUX clones (a-d) were performed but one representative experiment is shown.
