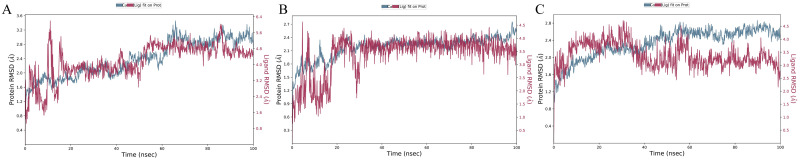
Fig 4. A 100-nanosecond simulation is conducted to measure the root mean square deviation (RMSD) results of three complexes.
Complexes 1, 3, and 6 are subjected to be a better binding stability over the 100-nanosecond molecular dynamics simulation using the Desmond software. (A) RMSD of Complex_1, (B) RMSD of Complex_3, and (C) RMSD of Complex_6. The root means square deviation (RMSD) between the ligand and protein exhibits temporal constancy, thereby ensuring stability. Nevertheless, complex_1 and 3 demonstrate persistent stability, suggesting that the interaction between the protein and ligand remains intact throughout the entire duration. Complex_6 exhibits a deviation of 30ns, indicating inferior stability compared to the other 2 complexes. Nevertheless, the overall binding interaction is not significantly unfavourable, and further investigation is required for the other parameters.