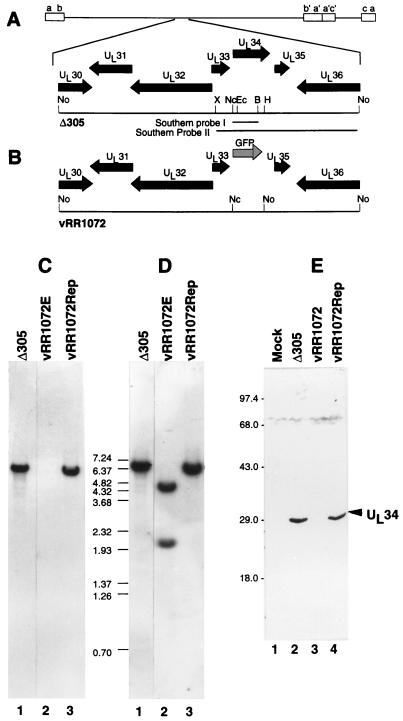
FIG. 2.
Structures of recombinant virus genomes and UL34 expression in recombinant viruses. (A and B) Sequence arrangement of viral genomes of Δ305 (A) and vRR1072 (B) around the UL34 locus, showing the positions of viral open reading frames (filled arrows), GFP insertion (shaded arrow), and probes used for Southern analysis of recombinant virus genome structure. Restriction enzyme abbreviations: B, BamHI; Ec, EcoRI; H, HpaI; Nc, NcoI; No, NotI; X, XbaI. (C) Autoradiographic image of a Southern blot of NotI-digested viral DNAs from cells infected with Δ305 (lane 1), the UL34 deletion virus vRR1072 (lane 2), or homologous repair virus vRR1072Rep probed with Southern probe I. (D) Same blot as in panel C but stripped and reprobed with Southern probe II. The sizes of migration standards (in kilobase pairs) are indicated between the panels. All lanes were from the same blot, but lane 1 was not adjacent to lanes 2 and 3 in the original autoradiogram. (E) Photographic image of a Western blot of SDS-PAGE-separated proteins from cells either mock infected (lane 1) or infected with Δ305 (lane 2), vRR1072 (lane 3), or vRR1072Rep (lane 4). The migration positions of size standards are indicated at the left. The position of the UL34 signal is indicated by the arrowhead.