Figure 3.
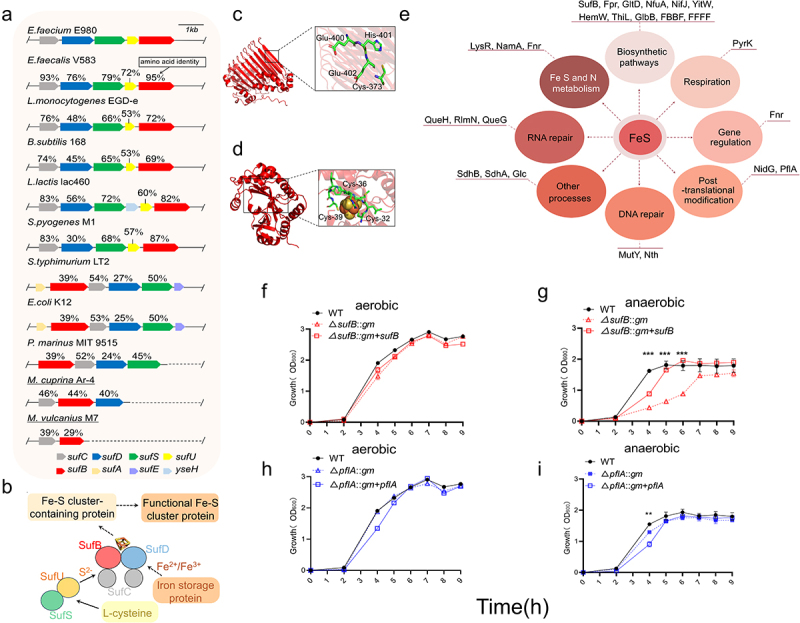
Phylogenetic and in silico analysis of Suf pathway.
(a) Phylogenetic analysis of suf gene cluster of E. faecium E980. The sufA, sufB, sufC, sufD, sufS, and sufU genes are color-coded to reflect their homology in different organisms. (b). Illustrations of Suf pathway for Fe-S cluster biogenesis in bacteria. Solid arrows denote functional steps that are amply corroborated by in vitro and in vivo data. The dashed arrows represent steps that have yet to be comprehensively characterized. The depicted surface representation structures include SufB (red), SufC (gray), SufD (blue), the SufS homodimer (green), and SufU (yellow). The putative Fe-S cluster binding sites of SufB (c) and PflA (d) in E. faecium. The specific residues needed to ligate Fe-S cluster are illustrated as sticks. (e) In silico analysis of Fe-S cluster-containing proteins in E. faecium E980 (This image was created in part By Figuredraw). Growth curves of E. faecium under both aerobic and anaerobic conditions. Growth curves of WT (black, solid line), ΔsufB::gm (red, dotted line), and ΔsufB::gm + sufB (red solid line) under both aerobic (f) and anaerobic (g) conditions. Similarly, growth curves of WT (black, solid line), ΔpflA::gm (blue, dotted line), and ΔpflA::gm + pflA (blue solid line) under both aerobic (h) and anaerobic (i) conditions were generated. The growth curves represent the average data from three independent experiments. Asterisks indicate significant differences between WT and mutant strains according to Student’s t test: *** P ≤ .001, ** P < .05.
