Figure 4.
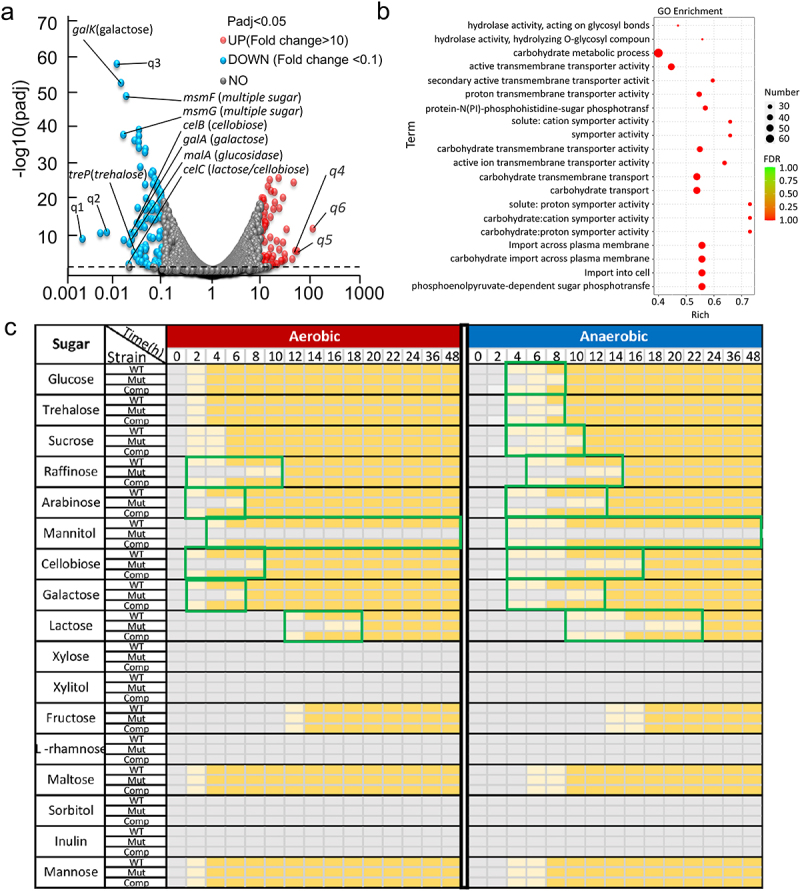
Transcriptome comparison between WT and sufB mutant strains under anaerobic conditions.
(a) Bubble plot of the DEGs (WT vs ΔsufB::gm). The dashed line indicates that P-value is equal to .05. The highly down-regulated and carbohydrate-related genes were labeled, in which the corresponding sugars are indicated in parentheses; q1-q6 represent the genes that were used for qPCR validation (Figure S6). (b) The top 20 enriched GO terms of the DEGs. (c) Bacterial strains (WT, ΔsufB::gm and ΔsufB::gm + sufB) were tested for their ability to metabolize 17 different sugars under aerobic and anaerobic conditions. The color gray and dark yellow represent negative and positive results for sugar metabolism, respectively. The light yellow represents intermediate result. The periods of metabolic gap between mutant and WT are marked in green boxes.
