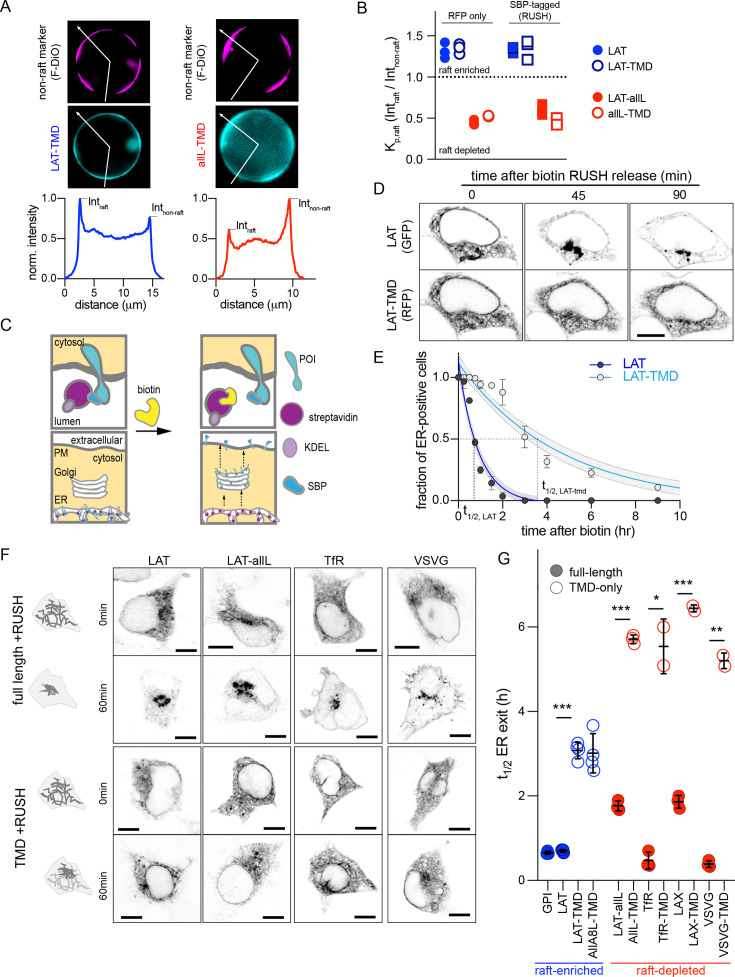
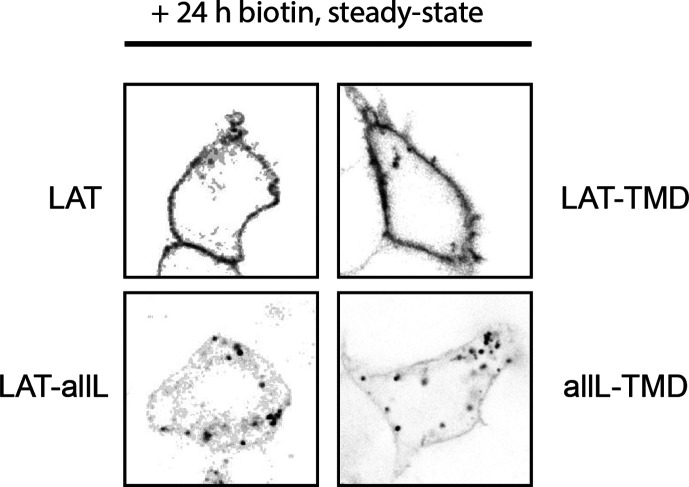
Figure 1. Full-length proteins exit the ER faster than TMD-only versions regardless of raft affinity.
(A) Exemplary images of GPMVs from cells expressing raft-preferring LAT-TMD (left) or non-raft-preferring allL-TMD (right). The RFP-tagged TMDs are shown in cyan; magenta shows the disordered phase marker Fast DiO (F-DiO). Bottom row shows fluorescence intensity line scans along white lines shown in cyan images, revealing protein partitioning between raft and non-raft phases. (B) The ratio of intensities in raft versus non-raft phase is the raft partition coefficient (Kp,raft). LAT-TMD and full-length LAT are enriched in the raft phase while allL-TMD (and full-length LAT with allL-TMD, LAT-allL) are largely depleted from raft phase. SBP-tagging (for RUSH assay) has no effect on raft affinity. Symbols represent 3 independent experiments with >10 GPMVs/experiment. All blue labeled constructs are not statistically different from one another, and each is P<0. 01 different from all red constructs. (C) Schematic of RUSH assay. (D) Confocal images of co-transfected LAT-EGFP and LAT-TMD-mRFP at various time points after biotin introduction. Full-length LAT exits ER faster. (E) Fraction of ER-positive cells decreases over time, allowing quantitative estimation of ER exit kinetics (t1/2). Symbols represent average +/-st.dev. from >3 independent experiments. Fits represent exponential decays with shading representing 95% confidence intervals. (F) Confocal images of various full-length and TMD-only RUSH constructs (RFP-tagged) at 0 and 60 min after biotin introduction. (G) Quantification of t1/2 for ER exit comparing full-length and TMD-only proteins (blue represents raft-enriched proteins, red = raft-depleted; see Supplementary file 1 for Kp,raft quantifications). Bars represent average ± st.dev. from three independent experiments; *p<0.05, **p<0.01, ***p<0.001. All scale bars correspond to 5 µm. Original data quantification can be found in the Source Data files.