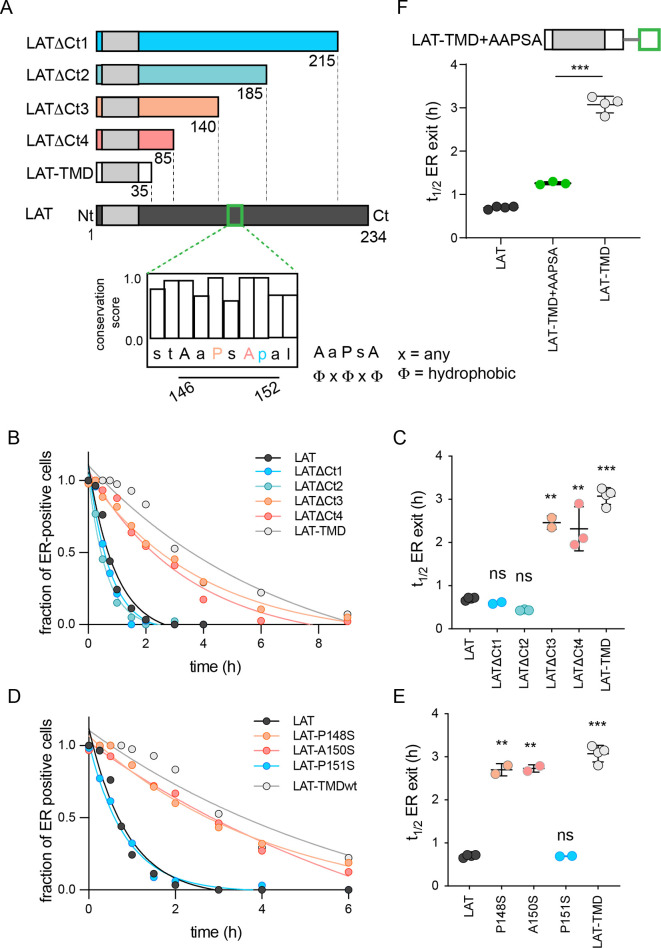
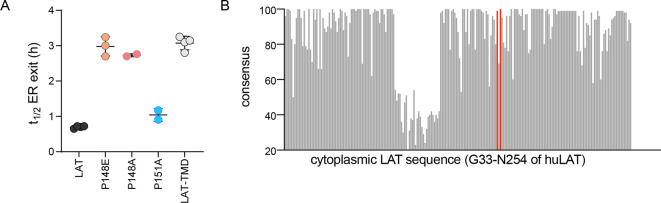
Figure 2. Identification of ER exit motif of LAT.
(A) Schematic of LAT and the truncated versions used here. Inset shows a putative COPII association motif and the evolutionary conservation of residues 144–153 of hLAT. (B) Temporal dependence of the fraction of ER-positive cells for LAT truncations. Deletion of a region comprised of residues 140–185 leads to slow ER exit. (C) Fitted ER exit kinetics for the constructs represented in panel C. Deletion of amino acids 140–185 slows ER exit kinetics by ~fourfold. (D) Temporal dependence of fraction of ER-positive cells with point mutations of ΦxΦxΦ motif. Mutations of key residues within the motif slow ER exit kinetics. (E) Fitted ER exit kinetics for point mutants in panel D. (F) Insertion of AaPsA motif into LAT-TMD accelerates ER exit kinetics. (B and D) show a representative experiment with exponential decay fits; points in C, E, and F represent t1/2 values of ER exit from fits of independent repeats with >20 cells/experiment. **p<0.01, ***p<0.001, nsp >0.05. Original data quantification can be found in the Source Data files.