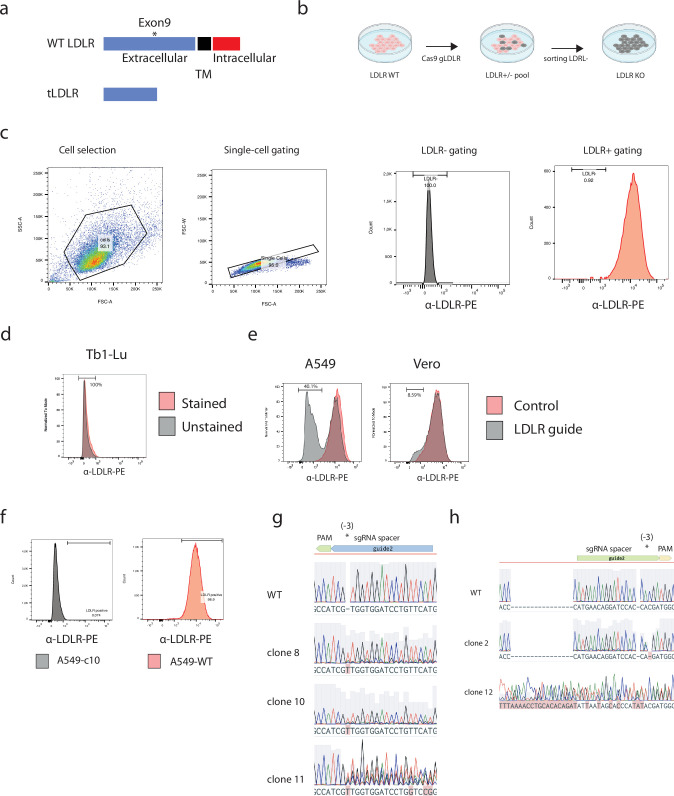
Extended Data Fig. 2. Generation and validation of knockouts in A549 and Vero cells.
a, Schematic of CRISPR-Cas9 editing strategy. The extracellular region of LDLR was targeted, leading to putative N-terminally truncated proteins not displayed on the cell surface for entry. b,Schematic of editing and α-LDLR sorting procedure. c, Gating strategy and PE intensity from α-LDLR-PE staining are shown. α-LDLR-PE staining was evaluated on single cells. Event densities were smoothened and are displayed as absolute counts or as counts normalization to the mode. Numbers indicate the percentage of single cells defined as α-LDLR-PE negative. d, Non-reactive and stained Tb1-Lu cells were used as negative control. PE intensity from α-LDLR-PE staining is shown. Event density was smoothened by normalization to the mode. e, Bulk sorting after transfection and transient Puromycin selection of α-LDLR-PE stained A549 or Vero cells, edited or unmodified (control) via CRISPR-Cas9. Event densities were smoothened and are displayed as counts normalization to the mode. f, Flow-cytometry result from A549-wild type and edited A549 clone 10 cells. PE intensity from α-LDLR-PE staining is shown. Event density was smoothened by normalization to the mode. Numbers indicate the percentage of single cells defined as α-LDLR-PE positive for A549 clone 10 and unmodified WT cells. Event densities were smoothened and are displayed as absolute counts. g, Sanger sequencing of PCR products from the LDLR genomic locus CRISPR-Cas9 editing site for A549 clones 8, 10 and 11 alongside unmodified wild-type cells are shown (via benchling.com alignment). The sgRNA spacer, PAM and expected Cas9 editing site (3 base-pairs downstream of PAM sequence) are shown above the sequencing traces. h, Same as shown for f, but for Vero cell clones C2, C12 and wild-type cells.