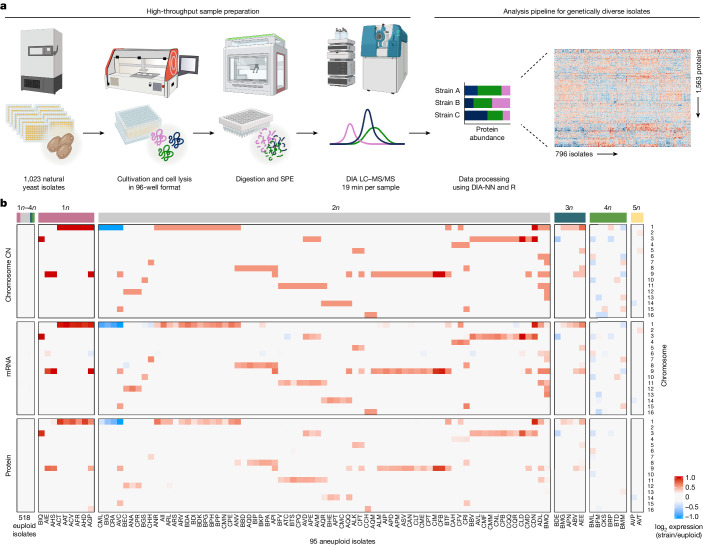
Fig. 1. High-throughput proteomics pipeline and assembly of a cross-omics dataset for studying aneuploidy in natural yeast isolates.
a, S. cerevisiae isolates were cultivated in synthetic minimal medium in 96-well format. Cells were collected by centrifugation at mid-log phase and lysed by bead beating under denaturing conditions. The lysate was then treated with reducing and alkylating reagents and digested with trypsin. The resulting peptides were desalted by solid-phase extraction (SPE) and analysed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) in data-independent acquisition (DIA) mode using SWATH-MS. Data were integrated using DIA-NN. b, Integrated dataset of 613 natural S. cerevisiae isolates. Relative chromosome copy number (CN), relative median mRNA levels and relative median protein abundances per chromosome between isolates and euploid reference (log2 ratios strain/euploid) are shown. The median across all euploid isolates was used as the reference for ratio calculations. Isolate ploidy is indicated at the top of the heat map.