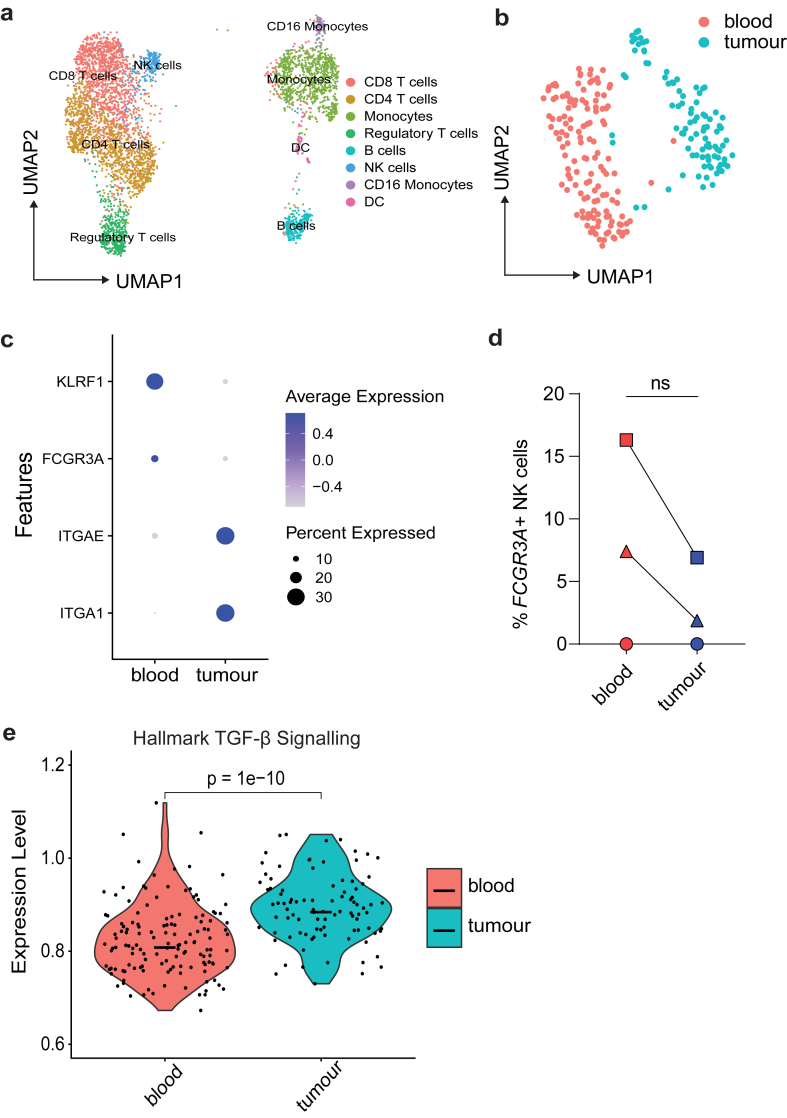
Fig. 1.
scRNA-seq of human bladder cancer identifies immunosuppressed tumour infiltrating NK cells. Bladder tumours harvested from patients and matched blood were analysed by scRNA-seq. (a) UMAP plot of CD45+ cells coloured clusters defined by canonical immune markers, where each dot represents a single cell. (b) UMAP plots of NK cell clusters where clusters separated by tissue compartment. (c) Dot plots showing expression of indicated genes across tumour or blood derived NK cell clusters. (d) Individual patient raw gene counts for indicated gene FCGR3A plotted as a percentage of total NK cells (n = 3). Data from one experiment (n = 3 biological samples) where each symbol represents an individual patient, graphs show mean value ± SEM. Groups were compared using Mann–Whitney t test, where P > 0.05 was deemed not significant. (e) Violin plots showing relative enrichment of indicated Hallmark gene sets, calculated by ssGSEA (n = 3). ssGSEA groups were compared using Welch's t test, where P > 0.05 was deemed not significant.