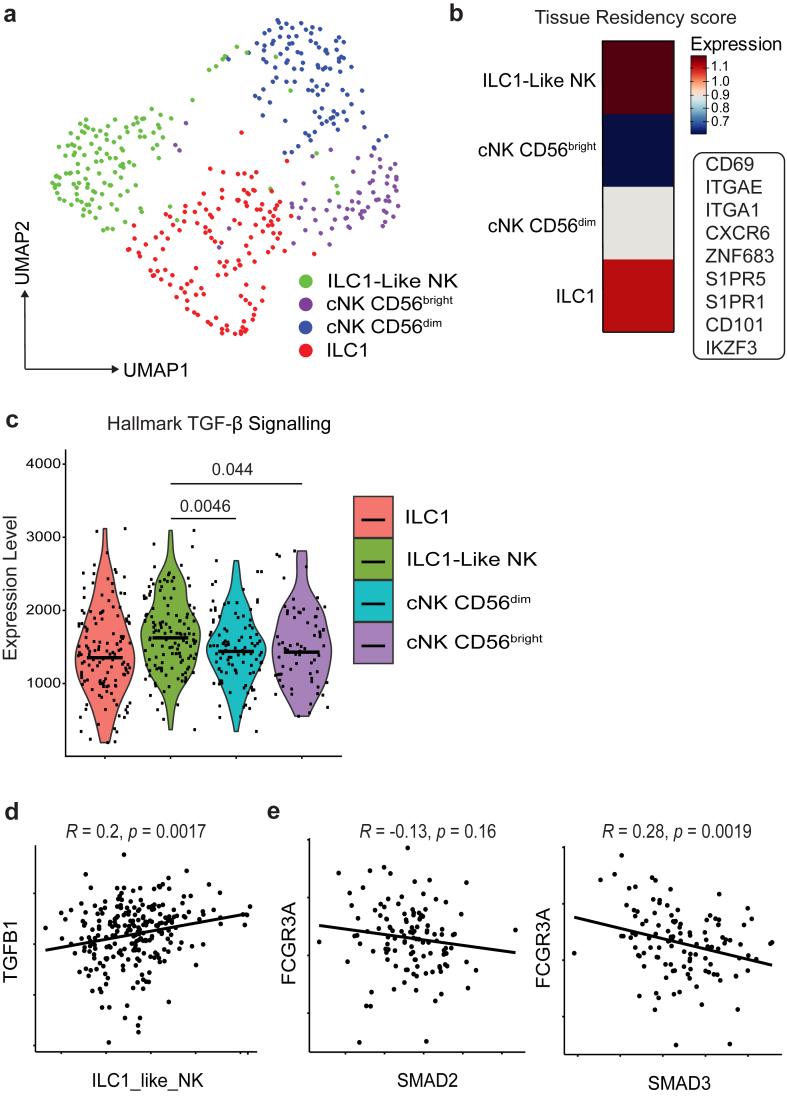
Fig. 3.
scRNA-seq and bulk sequencing datasets in bladder cancer further identifies TGF-β as a potential cause for CD16 loss. (a) UMAP plot showing NK cell clusters, integrated from 5scRNA-seq datasets from samples retrieved from patients with bladder cancer (n = 5). (b) Heatmap showing tissue residency score for each cluster as defined by indicated genes within the panel. (c) Violin plots showing relative enrichment of indicated Hallmark gene sets, calculated by ssGSEA. ssGSEA groups were compared using Welch's t test. (d) Correlation between ILC1-like NK score and TGFB1 subgroups in TCGA BLD cohort. (e) Correlation between FCRG3A and SMAD2/3 subgroups in TCGA BLD partitioned by high expression of NKscore. Coefficient was calculated with Pearson correlation analysis. Significant differences are indicated where P > 0.05 was deemed not significant.