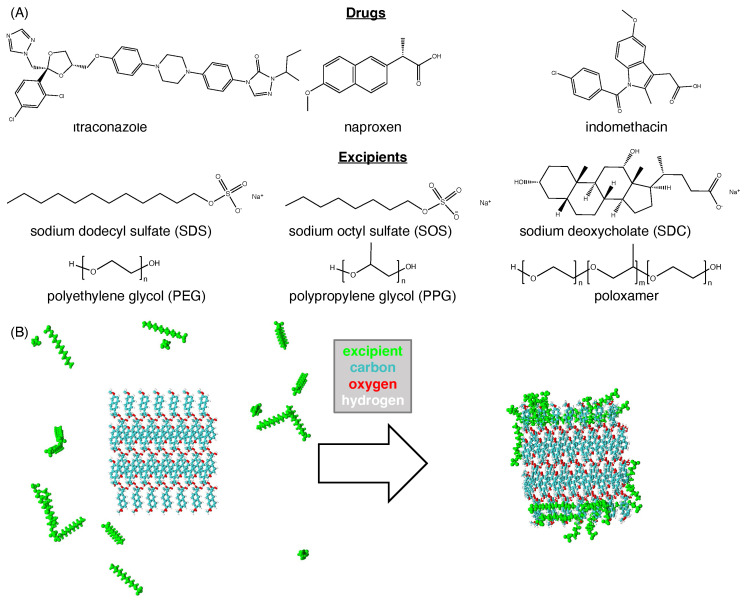
Figure 1.
Summary of the primary training dataset used in this study. (A) Molecular structures are shown for each of 3 drugs and 6 excipients. For each drug, each of the 6 excipients were independently simulated for a total of 18 different simulations. In simulations of PEG and PPG, n = 20. In simulations of poloxamer, n = 5 and m = 10, which gives the same length as the PEG and PPG used in the simulation. (B) An example of our simulation methodology, with images taken from the start (left) and end (right) of our simulation of a naproxen crystal with SDS. Water molecules are hidden for improved clarity. At the start, excipients (green) were placed randomly around the drug crystal (C–blue, O–red, and H–white). Over the course of the simulation, the majority of the excipients adsorb to the drug crystal.