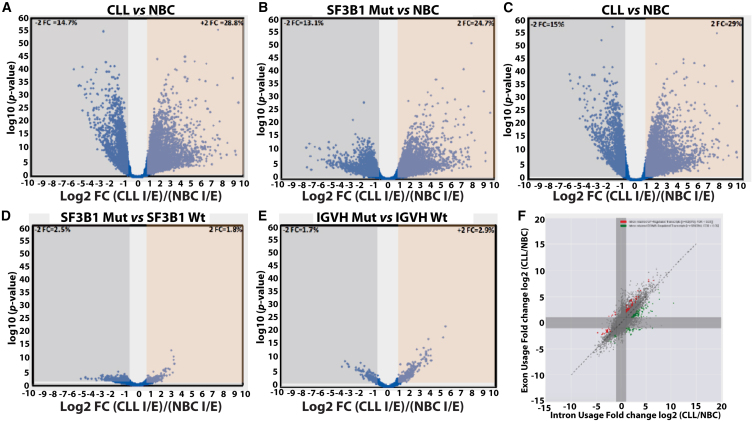
Figure 2.
RNA-sequencing analysis for global intron retention in CLL vs. normal B cells and correlation with transcript expression
RNA-seq analysis was conducted on sequences obtained from the EGA data derived from CLL samples (n = 97 including high risk = 41, low risk = 56) and normal B cells (n = 9 from healthy controls in triplicate). (A–E) Volcano plot displaying the differential intron retention log2 ratios TPM (intron/exon) in different combinations including (A) CLL-B cells and NBCs (CLL-B vs. NBC). The scatterplot shows 14811 transcripts for intron retention between CLL-B cells vs. NBCs. (B) CLL-B cells carrying Mut-SF3B1 and NBC (Mut-SF3B1 vs. NBC). (C) CLL-B cells carrying Wt-SF3B1 and NBC (Wt-SF3B1 vs. NBC). (D) CLL-B cells carrying Mut-SF3B1 and CLL-B cells carrying Wt-SF3B1 (Mut-SF3B1 vs. Wt-SF3B1). (E) CLL-B cells carrying Mut-IgVH and CLL-B cells carrying Wt-IgVH (Mut-IgVH vs. Wt-IgVH). The bottom black line represents the diagonal regression line where intron retention ratios are equal in both samples compared. (F) Further, the data were presented in form of a quadrant between CLL-B cells vs. NBCs to show the distribution of ratio of intronic and exonic TPM values for 14,811 transcripts.