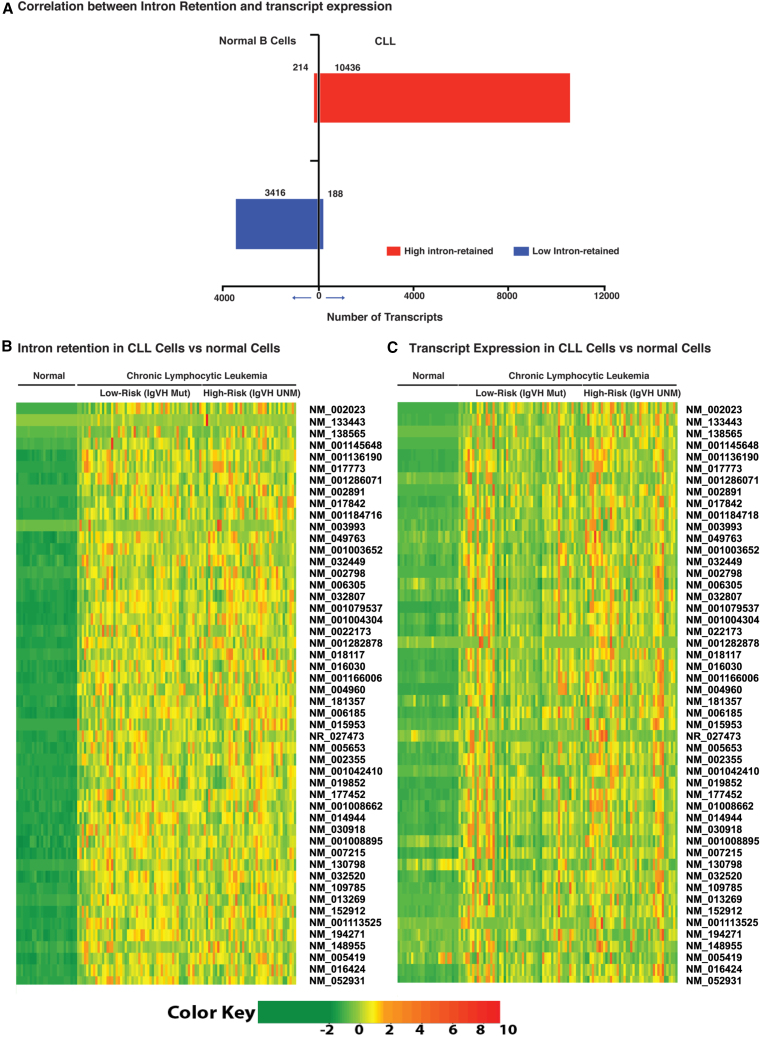
Figure 3.
The correlation between intron retention and transcript expression in various cell types
(A) A total of 16,725 transcripts that were accumulated differentially used-intron respectively in CLL (set-I) and NBC (set-II) at p ≤ 0.05 (FDR = 5%) and among those the three subsets each of the sets corresponding to over-expressed (set-IA, set-IIA), under-expressed (set-IB, set-IIB), and non-differentially expressed (set- IC, set-IIC) transcripts consequently found in CLL and NBC at p < 0.05 (FDR = 5%). The chi-square p value for the 2 × 3 table is ≤1.0e−300. The bar graph represents two sets of bars, each for a fraction of high intron used in CLL (set-I) and NBC (set-II) cases. It is associated with upregulated (set-IA, set-IIA) and downregulated (set-IB, set-IIB) at transcription level. The 2 × 2 contingency table shows significant p value <1.0e−300 and ODDs = 886. (B) The Heatmap Z scores of TPM values for sample-wise intron retention in NBC and CLL from each of the top 200 transcripts selected based on high intron retention in CLL with fulfilling condition CLL > NBC at p < 0.05; FDR = 5%. (C) The Heatmap Z scores of TPM values for sample-wise transcripts expression in NBC and CLL from each of the top 200 transcripts selected based on high intron retention in CLL with fulfilling condition CLL > NBC at p < 0.05; FDR = 5%.