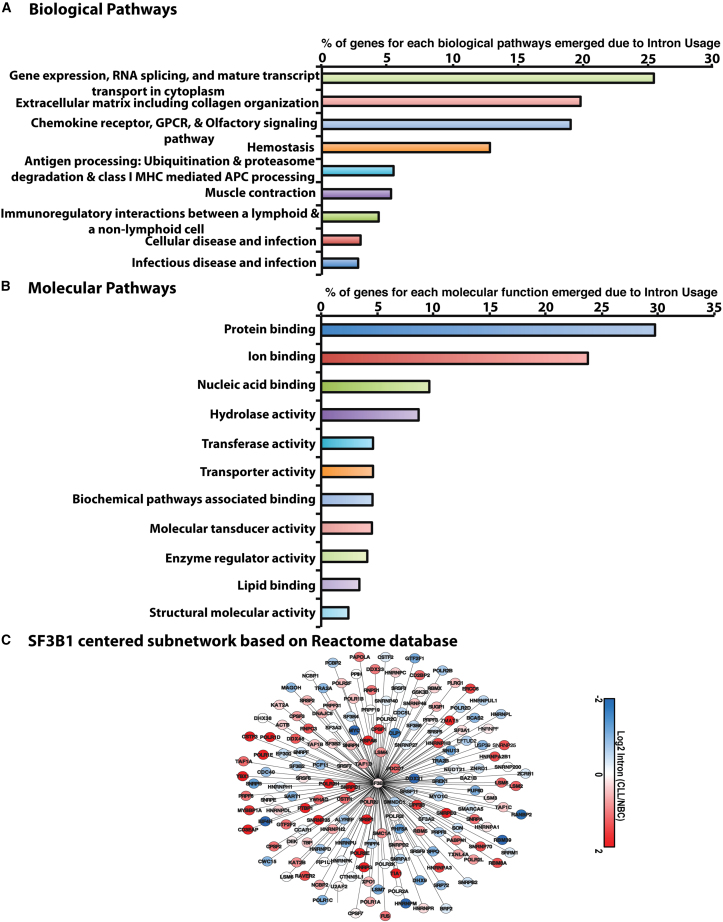
Figure 4.
Biological pathway, molecular functions, and network analysis based on intron retention
(A) For biological pathway analysis, we took the top 25% of transcripts with high intron retention in CLL as compared with normal B cells (p < 0.05, FDR = 0.1). The gene symbols for corresponding transcripts were used as input and subjected for Reactome Pathway. The p value was adjusted using a Bonferroni correction. The x axis shows the % of the genes enriched and y axis shows the description of the pathways. (B) Molecular function analysis was done for the top 25% of transcripts with high intron retention CLL/NBC Network analysis by selecting transcripts with high intron retention in CLL as compared with normal B cells using WebGestalt. The height of the bar represents the % of genes observed in the category. (C) Subnetwork centered on SF3B1 and its first neighbors. The SF3B1 centered subnetwork was based on Reactome FI network database and cytoscape V3.4 was used to identify the first neighbors of SF3B1. Nodes and links represent genes and functional interactions, respectively. The color of each node scale with log2 intron retention ratio in CLL cells vs. NBCs as indicated in the scale at the bottom.