Figure 5.
Selection and validation of transcripts for intron retention using RT-PCR
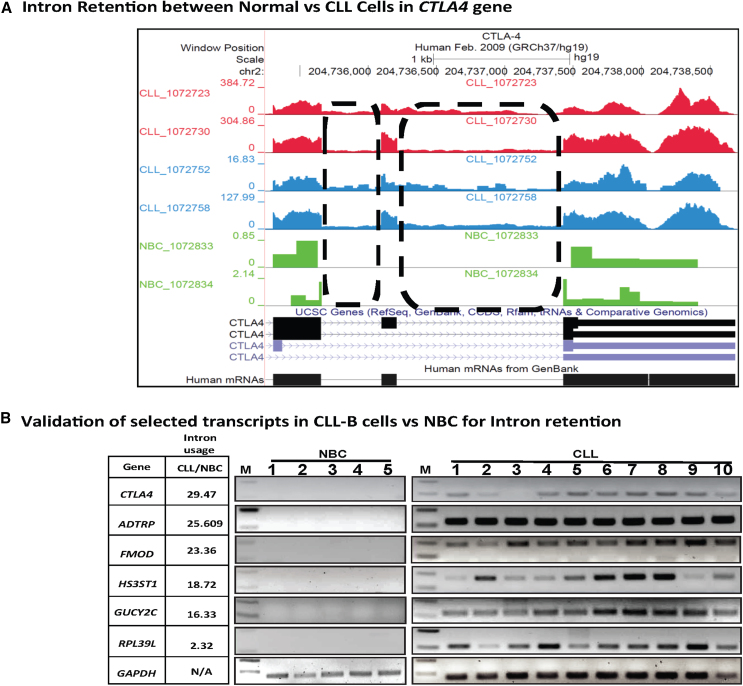
(A) RNA-seq read mapping (tracks) of the CTLA4 gene from the UCSC reference genome (hg19), six representative samples belonging to high- or low-risk CLL cells, or normal B cells are shown. The intron retention tracks are shown in green (for normal B cells), blue (low-risk CLL), and red (high-risk CLL) colors. It is clear that the reads map to the intronic region of CTLA4 transcript as annotated in the UCSC database. The absolute read counts for each sample are indicated on the y axis. (B) For validation, RNA was isolated from pure CLL-B cells or NBC and after DNase I digestion cDNA was prepared. Selected intronic region for assessment of intron retention were amplified using RT-PCR for PPP2R5B, ADTRP, RPL39L, HS3ST1, CTLA4, FMOD, and GUCY2C transcripts. GAPDH was used as control for normalization/loading of RNA.