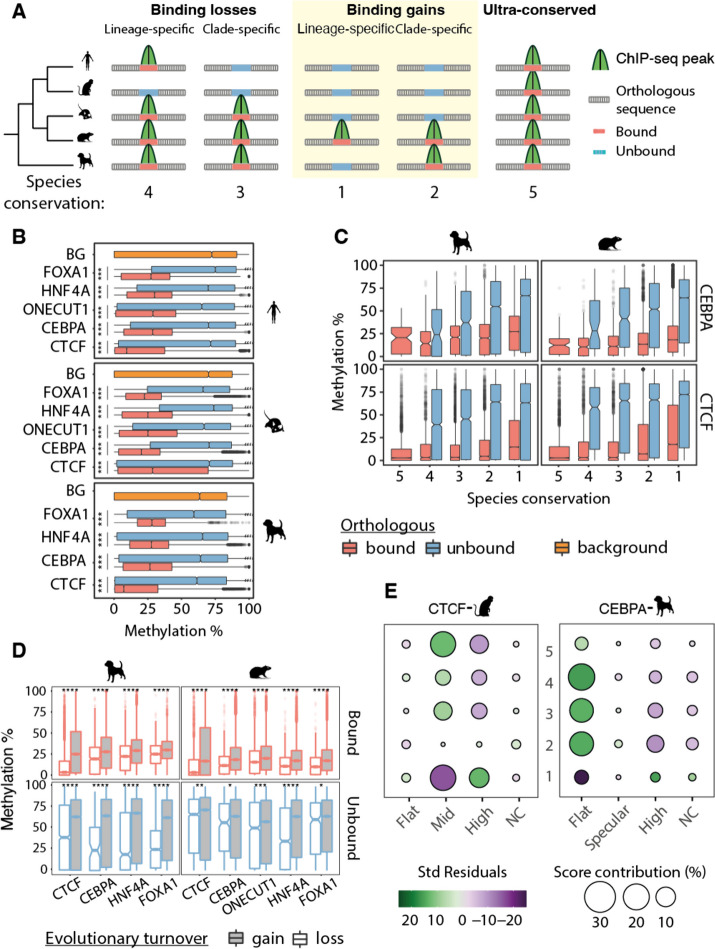
Fig. 4.
Coevolution of methylation and TF binding in mammals. A Schematic representation of the phylogenetic parsimony approach (adapted from [10]) to define species conservation categories and number of species with binding conservation. Briefly, TF binding events were first aligned and compared across species, then divided using parsimony in lineage- and clade-specific binding losses, and lineage- and clade-specific binding gains. Regions with experimentally determined binding in the species were called orthologous bound, and those without binding unbound. Ultra-conserved binding events were defined as those bound across all species. Below, examples of corresponding degrees of species conservation defined by the total number of species that share a TF binding event. B Average 5mC level distribution of orthologous bound regions, orthologous unbound regions, and genomic background (BG), with significant differences marked with asterixis (Wilcoxon test with Bonferroni correction, ***p-value ≤ 0.001). C Average 5mC distributions within CEBPA and CTCF orthologous bound and orthologous unbound regions divided by species conservation categories defined in panel A (Jonckheere-Terpstra trend test, p-values < 2.2e106), shown for dog and macaque. D Average 5mC distributions at orthologous bound and unbound regions of dog’s and rat’s TFBRs, further divided into evolutionary binding loss and gain events according to our parsimony approach. Orthologous sequences that concur in the definition of clade- or lineage-specific losses or gains are compared based on the presence (bound) or absence (unbound) of a binding event. Orthologous sequences defining a binding gain consistently have higher methylation levels than binding losses, both when unbound and bound by TFs (Wilcoxon test with Bonferroni correction, ***p-value ≤ 0.001). E Relationships between species conservation and 5mC profiles. Balloon plots show standardized residuals from an association analysis (chi-square test of independence) between 5mC profiles and TF binding conservation categories for dog’s CEBPA and macaque’s CTCF TF binding events. Positive residuals indicate a positive association between the degree of species conservation and methylation profile, while negative residuals indicate negative associations. For example, dog’s CEBPA binding events with a flat profile are positively associated with higher levels of species conservation, while they are negatively associated with lineage-specific binding events. The size of the balloons is proportional to the percentage of contribution to the total Chi-square score, therefore highlighting the most influencing combination of species conservation and methylation profile to the overall statistics