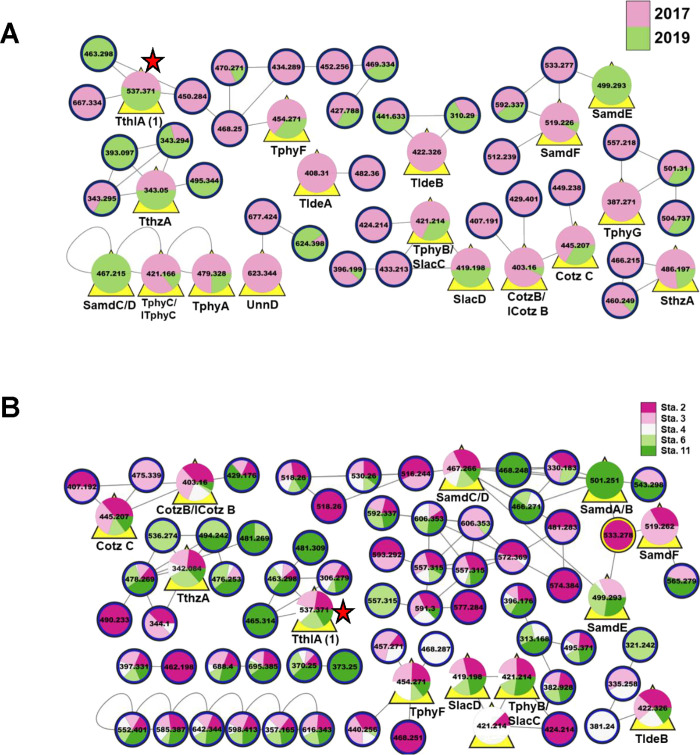
Figure 1.
T. thiebautii specialized metabolites were found year after year and across the GoM. (A) Partial LC-MS/MS-based molecular network of extracts from collection GoM2017 (pink) and GoM2019-6 (green). (B) T. thiebautii specialized metabolites found across the GoM in 2019. Partial LC-MS/MS-based molecular network of extracts from 2019 collections (stations 2–4, 6, and 11). In all panels, nodes are designated by their precursor masses. In panels (A) and (B), yellow triangles behind the node indicate that these metabolites were previously characterized by our group (cf. Figure S4). In panel (B), “pie slices” designate stations where the metabolite was detected. Red stars show the position of the node for trichothilone A (1) (m/z 537). Cotz, conulothiazole; ICotz, isoconulothiazole; ITphy, isotrichophycin; Sam, smenamide; Slac, smenolactone; Sthz, smenothiazole; Tlde, tricholide; Tphy, trichophycin; Tthl, trichothilone; Tthz, trichothiazole; Unn, unnarmicin. The full networks can be found at http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task = 27cc1b6493804e0283a08d5d056d015d (temporal–total number of nodes in full network 1465), http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4b5821e25366435fae44da433db8819e (spatial—total number of nodes in full network 538).