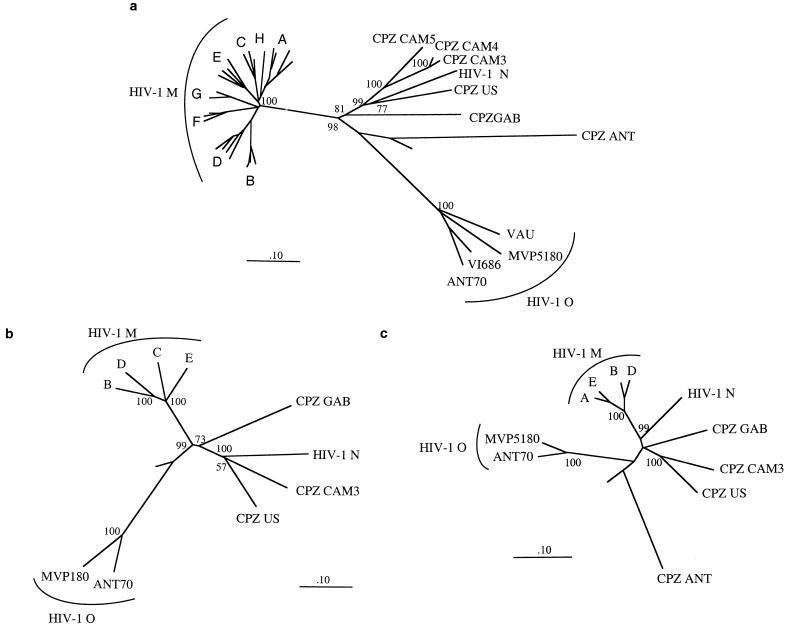
FIG. 1.
Phylogenetic relationship of SIVcpz-cam with other members of the HIV-1 and SIVcpz lineage. Nucleotide sequences for all viruses but SIVcpz from Cameroon were obtained from the HIV data bank (http://hiv-web.lanl.gov/). HIV-1 N is represented by the YBF30 strain (23). Sites with gaps or unidentified nucleotides were stripped from the alignment. The trees were rooted with HIV-2 Rod. Trees based on the codon-based nucleotide alignments and constructed by the neighbor-joining method are shown. The percentages of concordant branching during bootstrapping in 10,000 resamplings are displayed at the branch nodes. The branch lengths are drawn to scale. The bar indicates a genetic distance of 0.1 (10% of nucleotide divergence). The neighbor-joining trees were consistent with those inferred by the Fitch-Margoliash, the maximum parsimony, and the maximum likelihood methods. Only minor variations in branch lengths were seen for the maximum likelihood trees. No significant differences in the trees based on the amino acids of Pol and Env were observed. (a) Phylogenetic relationship of SIVcpz-cam3, SIVcpz-cam4, and SIVcpz-cam5 with other members of the HIV-1 and SIVcpz lineage in a partial (1,332-bp) env fragment (from C2 to the ectodomain of gp41); (b) SIVcpz-cam3 phylogenetic relationships with other HIV-1 and SIVcpz viruses in the whole env gene (2,164 nucleotides included); (c) SIVcpz-cam3 phylogenetic relationships with other HIV-1 and SIVcpz viruses in the pol gene.