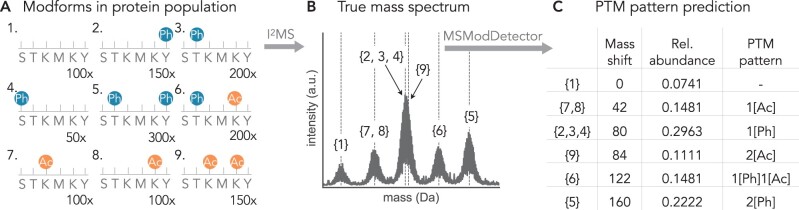
Figure 1.
PTM pattern inference problem. The protein population in (A) can be modified by phosphorylation (Ph) at the serine (S), threonine (T), or tyrosine (Y) sites or it can be acetylated (Ac) at the lysine (K) sites, leading to different combinations of PTMs (“modforms”) within the population of the same protein. All 32 possible modforms are listed in Supplementary Fig. S1. The numbers on the bottom right indicate the amount of the respective modform. (B) I2MS measures the total mass of each protein molecule and generates a true mass spectrum. Modforms 2, 3, and 4 have the same mass shift and the same isotopic distribution, as do modforms 7 and 8. These “patterns,” which have the same numbers of different modifications, are what is resolved by I2MS. The patterns with one phosphorylation (modforms 2, 3, 4) or 2 acetylations (modform 9) cannot be resolved visually in the spectrum as their isotopic distributions overlap too closely. (C) The goal of MSModDetector is to identify mass shifts caused by the modifications and to infer which PTM patterns are present and estimate their abundance using linear programming.