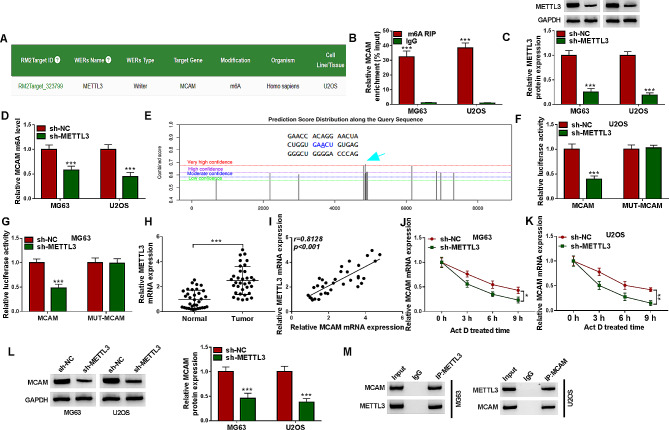
Fig. 4.
METTL3 targeted MCAM. (A) The m6A methylation modification sites of METTL3 in MCAM was predicted by RM2target database. (B) After MeRIP, MCAM enrichment was determined by qRT-PCR. (C) The protein level of METTL3 in MG63 and U2OS cells transfected with sh-NC or sh-METTL3 was measured via western blot. (D) MCAM m6A level in MG63 and U2OS cells transfected with sh-NC or sh-METTL3 was determined. (E) SRAMP predicted MCAM mRNA contained the m6A modification site of METTL1. (F and G) The interaction between MCAM and METTL3 was verified by dual-luciferase reporter assay. (H) The mRNA expression of METTL3 in OS tissues and normal tissues was determined by qRT-PCR. (I) The linear correlation between METTL3 mRNA level and MCAM mRNA level in OS tissues was estimated by Spearman’s coVrrelation coefficient analysis. (J and K) After treatment with Act D for 0 h, 3 h, 6 h and 9 h, MCAM mRNA expression was determined by qRT-PCR. (L) MCAM protein level in MG63 and U2OS cells with sh-NC or sh-METTL3 transfection was measured via western blot. (M) The binding association between MCAM and METTL3 was analyzed by Co-IP assay. *P < 0.05, **P < 0.01, ***P < 0.001