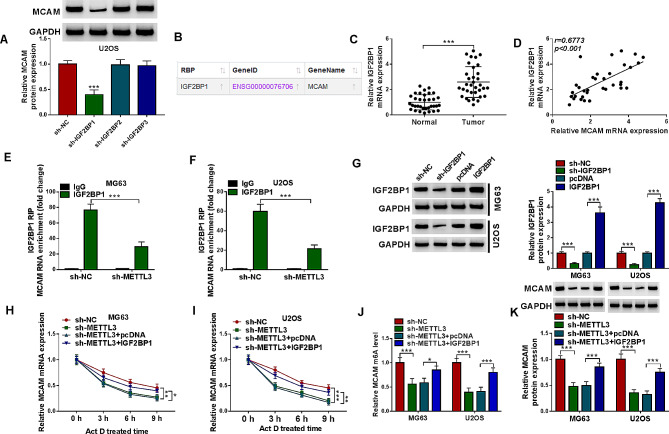
Fig. 5.
IGF2BP1 affected METTL3-mediated the m6A methylation modification of MCAM. (A) After sh-NC, sh-IGF2BP1, sh-IGF2BP2 or sh-IGF2BP3 transfection in MG63 and U2OS cells, MCAM protein level was measured by western blot. (B) The relation between IGF2BP1 and MCAM was predicted by ENCORI. (C) IGF2BP1 mRNA level in OS tissues and normal tissues was determined by qRT-PCT. (D) The linear correlation between the mRNA levels of IGF2BP1 and MCAM in OS tissues was analyzed. (E and F) The effect of METTL3 knockdown on the interaction between IFG2BP1 and MCAM was estimated by RIP assay. (G) The protein level of IGF2BP1 in MG63 and U2OS cells transfected with sh-NC, sh-IGF2BP1, pcDNA or IGF2BP1 was measured by western blot. (H and I) After Act D treatment, MCAM mRNA level in MG63 and U2OS cells transfected with sh-NC, sh-METTL3, sh-METTL3 + pcDNA or sh-METTL3 + IGF2BP1 was determined by qRT-PCR. (J) The effects of METTL3 and IGF2BP1 on MCAM m6A level in MG63 and U2OS cells transfected with sh-NC, sh-METTL3, sh-METTL3 + pcDNA or sh-METTL3 + IGF2BP1 were analyzed. (K) MCAM protein level in MG63 and U2OS cells transfected with sh-NC, sh-METTL3, sh-METTL3 + pcDNA or sh-METTL3 + IGF2BP1 was measured by western blot. *P < 0.05, **P < 0.01, ***P < 0.001