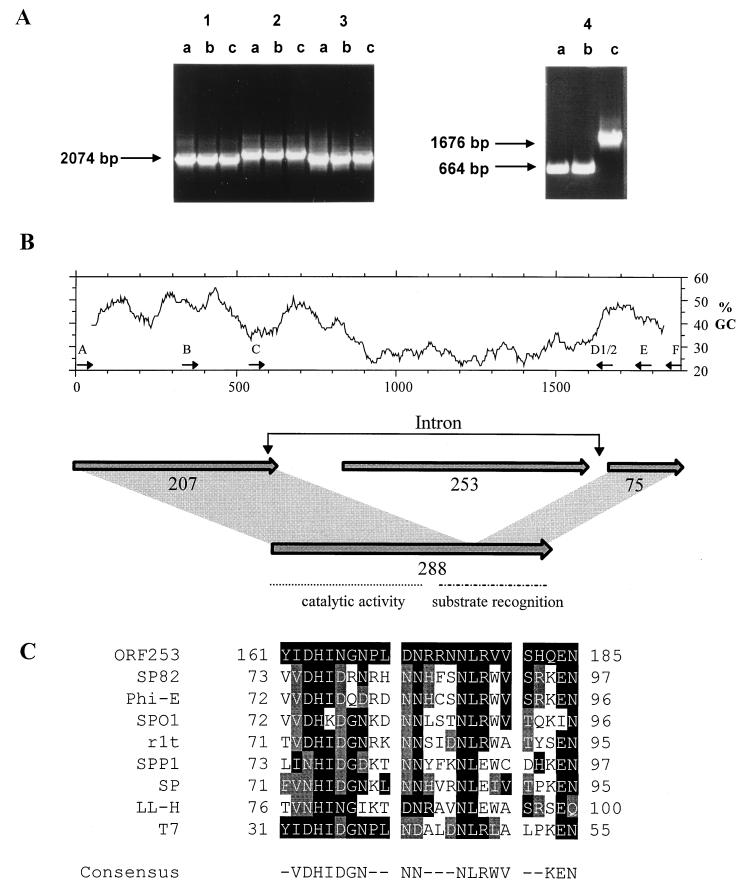
FIG. 1.
(A) PCR screening of S. thermophilus bacteriophage genomes for insertion or deletion events. PCR amplification products obtained with primer pairs G1-G2 (block 1), H1-H2 (block 2), I1-I2 (block 3), and A-E (block 4) and phage DNAs from Sfi21 (lanes a), S3 (lanes b), and S3b (lanes c) are shown. Molecular sizes of the PCR products are provided for blocks 1 and 4. (B) (Top) Percent GC content distribution of the investigated 1.9-kb region of phage S3b encompassing the lysin gene (window length, 100 bp). Primers referred to throughout the text are indicated by thin arrows above the ruler providing the nucleotide scale in base pairs. (Bottom) Prediction of open reading frames in the region of the phage S3b lysin gene. The open reading frames are indicated by shaded arrows, with their lengths given in amino acids. orf 288, which results from an orf 207-orf 75 fusion, is shown together with the domain structure deduced from knowledge of pneumococcal phage Dp-1 (33). (C) Multiple alignment of a 25-aa block from the S3b orf 253 gene product and the gene product of coliphage T7 gene 3.8 (V01146), the gene product of Bacillus phage SPP1 orf 36.1 (X67865), and the intron-encoded gene products of Bacillus phages SP82 (U04812), phi-E (U04813), SPO1 (P34081), and SP (L31962), L. lactis phage r1t (U38906), and Lactobacillus phage LL-H (L37351). Amino acid positions identical to those of S3b are shaded in black, while conserved amino acids are shaded in grey. The consensus sequence indicates identical amino acids present in five or more sequences.