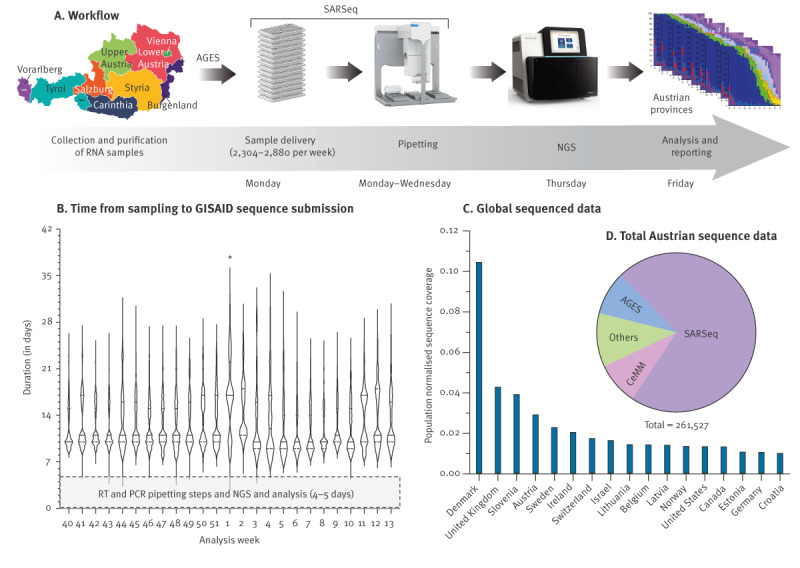
Figure 1.
(A) Workflow and (B,C) efficiency of the Austrian SARS-CoV-2 genomic surveillance, as well as (D) relative contribution of SARSeq to the surveillance system, Austria, January 2021–March 2023
AGES: Austrian Agency of Health and Food Safety; CeMM: Research Center for Molecular Medicine of the Austrian Academy of Sciences; IMBA: Institute of Molecular Biotechnology of the Austrian Academy of Science; NGS: next generation sequencing; RT: reverse transcription.
A. Workflow of the Austrian genomic surveillance pipeline. RNAs from individuals are extracted by laboratories in the country’s provinces and AGES selects representative samples, which are delivered to IMBA where they are processed through SARSeq (pipetting for RT, PCRs) in preparation for NGS. After NGS, semi-automated analysis of sequence data enables reporting of the results.
B. Violin plots showing the duration in days from sample collection to sequence submission to GISAID per analysis week. The grey dotted box highlights the duration of RT, PCR, NGS and analysis. The asterisk (*) denotes a holiday-related analysis week, resulting in older processed samples due to pipeline inactivity.
C. Sequenced data obtained by Austria and other countries, in the period of SARSeq pipeline operation. Data are normalised to country population. Coverage was determined using information on amounts of generated sequences, available from GISAID (from the SARSeq period: 01.2021 to 03.2023, accessed April 2023), and population counts from Our World in Data (https://ourworldindata.org/grapher/population?time=latest; accessed April 2023).
D. Total sequenced data originating from Austria during the SARSeq pipeline operation.