Figure 3.
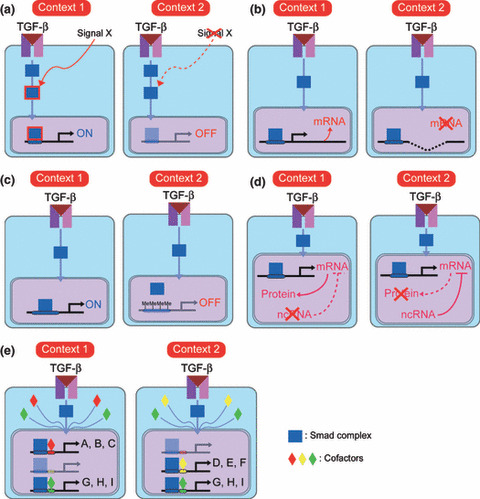
Several mechanisms of cellular context‐specific diversity of transforming growth factor‐β (TGF‐β)‐induced cell responses. (a) “Signal cross‐talk” model. In “context 1”, but not “context 2”, signal X is transduced in cells to modify downstream transducers of TGF‐β signaling and induce a certain “context 1”‐specific cell response. (b) “Genetic alterations” model. In “context 1”, expression of a certain target gene is induced by TGF‐β signaling. In “context 2”, the gene is deleted at the chromosomal level and TGF‐β stimulation fails to induce its expression. (c) “Epigenetics” model. In “context 1”, promoter regions of certain TGF‐β target genes adopt an “open conformation” and are exposed to the Smad complex. Conversely, in “context 2”, promoter regions of the same target genes adopt a “closed conformation” and the Smad complex fails to access the Smad‐binding elements. This difference results in differential responses to TGF‐β stimulation. (d) “Non‐coding RNA” model. In “context 2”, transcribed mRNAs of TGF‐β target genes are negatively regulated by non‐coding RNA (ncRNA). In “context 1”, such ncRNA is not expressed, resulting in translation of the mRNAs. (e) “Cofactors” model. Almost all TGF‐β target genes (A–I) are regulated by Smad proteins. Expression profiles of cofactors of Smad proteins differ between “context 1” and “context 2”, resulting in different responses to TGF‐β stimulation.
