Abstract
To identify novel cancer‐promoting genes in biliary tract cancer (BTC), we constructed a retroviral cDNA expression library from a clinical specimen of BTC with anomalous pancreaticobiliary duct junction (APBDJ), and used the library for a focus formation assay with 3T3 fibroblasts. One of the cDNAs rescued from transformed foci was found to encode Indian hedgehog homolog (IHH). The oncogenic potential of IHH was confirmed both in vitro with the focus formation assay and in vivo with a tumorigenicity assay in nude mice. The isolated IHH cDNA had no sequence alterations, suggesting that upregulation of IHH expression may contribute to malignant transformation. Quantitation of IHH mRNA among clinical specimens has revealed that the expression level of IHH in BTC with APBDJ is higher than that in BTC without APBDJ and than in non‐cancerous biliary tissues. Our data thus implicate a direct role of IHH in the carcinogenesis of BTC with APBDJ. (Cancer Sci 2009)
Biliary tract cancer (BTC) is a highly fatal malignancy in humans, and is prevalent in South American and Asian countries; approximately sixteen thousand people die of BTC every year in Japan.( 1 ) Unfortunately, many BTC cases are diagnosed at advanced clinical stages with a 5‐year survival rate of ∼10%.( 2 , 3 , 4 ) Several risk factors for BTC have been identified to date, including cholelithiasis,( 5 ) anomalous pancreaticobiliary duct junction (APBDJ),( 6 ) and primary sclerosing cholangitis.( 7 ) Genetic alternations in KRAS or TP53 and/or overexpression of ERBB2 have been shown to contribute to the development of certain types of BTC. However, many cases with BTC do not harbor any such genetic changes, and other transforming events further await discovery.
The focus formation assay with 3T3 or RAT1 fibroblasts has been extensively used to screen for transforming genes in various carcinomas.( 8 ) In such screening, genomic DNA is isolated from cancer specimens, and used to transfect 3T3 fibroblasts to obtain transformed cell foci. As expression of transfected genes in 3T3 cells in this assay is regulated by their own promoter and enhancer fragments, oncogenes with tissue‐specific expression (e.g. those with a blood cell‐specific promoter) can not become transcriptionally active in 3T3 cells, and thus can no longer be captured in such a screening system.
To ensure the sufficient expression of oncogenes in 3T3 cells, their transcription should be directly regulated by an exogenous promoter fragment. We have therefore constructed a retroviral cDNA expression library from a surgically operated clinical specimen of BTC with APBDJ, which was subsequently used to infect 3T3 cells. In the preparation of the cDNA library, we further took advantage of the SMART PCR system (Clontech, Mountain View, CA, USA), which preferentially amplifies full‐length cDNA. A focus formation assay with the library has resulted in the identification of a transforming Indian hedgehog homolog (IHH) cDNA.
Materials and Methods
Focus formation assay with a retroviral library. A recombinant retroviral library was constructed as described previously,( 9 , 10 , 11 , 12 ) with minor modifications. In brief, total RNA was extracted from a BTC specimen with APBDJ isolated from a 67‐year‐old man, who gave informed consent. This study was approved by the ethics committee of Jichi Medical University. First‐strand cDNA was synthesized from the RNA with the use of PowerScript reverse transcriptase, the SMART IIA oligonucleotide, and CDS primer IIA (all from Clontech). The resulting cDNA was then amplified by PCR with 5′‐PCR primer IIA (Clontech) and PrimeSTAR HS DNA polymerase (Takara Bio, Shiga, Japan) for 18 cycles of 98°C for 10 s and 68°C for 6 min. The PCR products were ligated to a BstXI adapter (Invitrogen, Carlsbad, CA, USA) and then incorporated into the pMXS retroviral plasmid (kindly provided by T. Kitamura of the Institute of Medical Science, University of Tokyo). A total of 5.8 × 105 colony forming units of independent plasmid clones was thus generated. Twenty clones were randomly isolated from the library, and examined for the incorporated cDNA. Sixteen (80%) out of the 20 clones contained cDNA inserts with an average length of 1.16 kbp. Recombinant retroviruses were produced by introduction of the plasmid library into the packaging cell line BOSC23 (American Type Culture Collection, Manassas, VA, USA) and were used to infect 3T3 cells in the presence of 4 μg/mL polybrene (Sigma, St Louis, MO, USA). The cells were cultured for 2 weeks, after which transformed foci were isolated, expanded, and subjected to extraction of genomic DNA. Insert cDNA was recovered from the genomic DNA by PCR with 5′‐PCR primer IIA and PrimeSTAR HS DNA polymerase. Amplified products were then ligated to the plasmid pT7Blue‐2 (Novagen, Madison, WI, USA) and subjected to nucleotide sequencing.
Tumorigenicity assay in nude mice. 3T3 cells (2 × 106) were infected with a retrovirus expressing IHH, resuspended in 500 μL PBS, and injected into each shoulder of a nu/nu Balb‐c mouse (6 weeks old). Tumor formation was assessed after 2 weeks.
Anchorage‐independent growth in soft agar. 3T3 cells (2 × 106) were infected with a retrovirus encoding IHH or v‐Ras, resuspended in the culture medium supplemented with 0.4% agar (Sea Plaque GTG agarose; Cambrex, East Rutherford, NJ, USA), and seeded onto a base layer of complete medium supplemented with 0.5% agar. Cell growth was assessed after culture for 2–3 weeks.
Quantitative RT‐PCR analysis. Portions of oligo(dT)‐primed cDNA produced by reverse transcription were subjected to PCR with a QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA, USA) and an amplification protocol comprising incubation at 94°C for 15 s, 60°C for 30 s, and 72°C for 60 s. Incorporation of the SYBR Green dye into PCR products was monitored in real time with an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA), thereby allowing determination of the threshold cycle (C T) at which exponential amplification of PCR products begins. The C T values for cDNA corresponding to the β‐actin gene (ACTB) and IHH were used to calculate the abundance of the latter mRNA relative to that of the former. The oligonucleotide primers used for PCR were 5′‐CCATCATGAAGTGTGACGTGG‐3′ and 5′‐GTCCGCCTAGAAGCATTTGCG‐3′ for ACTB and 5′‐CCTCTCTCCTAGAGACCTTG‐3′ and 5′‐CTGGCTCCCAGGGAATTTAG‐3′ for IHH.
Immunohistochemistry. Human tissues were fixed in 4% formaldehyde in PBS overnight at room temperature, embedded in paraffin, and sectioned at a thickness of 3 μm. Sections were mounted on glass slides, deparaffinized in three changes of xylene for 4 min each, and rehydrated in distilled water through a series of graded alcohols. For histological evaluation, sections were stained with hematoxylin–eosin. For immunohistochemical experiments, antigenicity was enhanced by boiling the sections in 10 mm citrate buffer (pH 6.0) in a microwave oven for 15 min, and the endogenous peroxidase activity was blocked by incubation in methanol containing 0.3% H2O2 for 30 min. After two washes with PBS containing 1% Triton X‐100, the sections were preincubated with the blocking buffer (#X0909; Dako, Glostrup, Denmark) in a humidified chamber for 20 min at room temperature, and then incubated overnight at 4°C with anti‐IHH antibody (sc‐1196; Santa Cruz Biochemistry, Santa Cruz, CA, USA) diluted in PBS. Next, the sections were washed in PBS and incubated with horseradish peroxidase‐labeled polymers conjugated to secondary antibodies for primary rabbit antigoat immunoglobulin (Dako, #P0449) without dilution at 37°C for 30 min. Color development was carried out by incubating the sections with 3,3‐diaminobenzidine tetrahydrochloride (Wako Pure Chemical Industries, Osaka, Japan) as the chromogenic substrate. Finally, the sections were lightly counterstained with hematoxylin, mounted, and viewed under a light microscope. For the negative control, the immunostaining processes were carried out by replacing the primary antibody with PBS.
Results
Screening with the focus formation assay. From the mRNA of a BTC specimen with APBDJ, full‐length cDNA was selectively amplified and ligated to a retroviral vector pMXS. From such library plasmids, we generated a recombinant ecotropic retrovirus that was subsequently used to infect mouse 3T3 fibroblasts. Infection experiments were repeated for a total of four times. After 3 weeks of culture, 75 transformed foci were observed. No foci could be found among the cells infected with an empty virus, while numerous foci were easily identified in the cells infected with a virus expressing v‐Ras oncoprotein (data not shown).
Each focus was isolated, expanded independently, and used to prepare genomic DNA. We then tried to recover retroviral inserts from such genomic DNA by PCR amplification with the primer used originally to amplify the cDNA in the construction of the library. In most cases, one to three DNA fragments were recovered from each genome, implying multiple retroviral infection of some 3T3 cells.
We finally obtained a total of 44 cDNA fragments by PCR, each of which was ligated into a cloning vector, and subjected to nucleotide sequencing from both ends. Screening of the 44 cDNA sequences against the public nucleotide sequence databases revealed that the 44 fragments correspond to 29 independent genes (Table 1).
Table 1.
Bile duct cancer cDNA isolated from 3T3 transformants
| Clone ID # | Gene symbol | GenBank no. | Presence of full ORF |
|---|---|---|---|
| 1 | FAM83H | NM_198488 | No |
| 2 | GATAD1 | NM_021167 | Yes |
| 3 | RRAS2 | NM_012250 | No |
| 4 | FASTK | NM_006712 | Yes |
| 5 | VAT1 | NM_006373 | Yes |
| 6 | ARPC2 | NM_005731 | No |
| 7 | IHH | NM_002181 | Yes |
| 8 | SENP6 | NM_015571 | Yes |
| 9 | DOT1L | NM_032482 | ND |
| 10 | LTBR | NM_002342 | ND |
| 11 | KRAS | NM_004985 | Yes |
| 12 | TMEM54 | NM_033504 | Yes |
| 13 | RNASET2 | NM_003730 | Yes |
| 14 | RPS4X | NM_001007 | Yes |
| 15 | TETRAN | NM_001120 | Yes |
| 16 | DFNB31 | NM_015404 | No |
| 17 | CLDN3 | NM_001306 | No |
| 18 | GJB2 | NM_004004 | Yes |
| 19 | PSMA7 | NM_002792 | Yes |
| 20 | PRPSAP1 | NM_002766 | Yes |
| 21 | LRRC59 | NM_018509 | Yes |
| 22 | LRP5 | NM_002335 | ND |
| 23 | NCOR2 | NM_006312 | No |
| 24 | KLF16 | NM_031918 | No |
| 25 | ARHGAP4 | NM_001666 | ND |
| 26 | KIAA0284 | NM_015005 | No |
| 27 | DNAJC4 | NM_005528 | ND |
| 28 | NOTCH2NL | NM_203458 | No |
| 29 | BCKDHB | NM_000056 | Yes |
ND, not determined; ORF, open reading frame.
Identification of IHH. To confirm the transforming potential of the isolated cDNA, each cDNA clone was ligated to pMXS, and corresponding retrovirus was used to re‐infect 3T3 cells. Focus formation assays were conducted for 13 independent genes, discovering a reproducible transforming activity for clone ID #7 corresponding to IHH (GenBank accession number, NM_002181) (Fig. 1, top panel). Again, infection with a virus for v‐Ras induced many transformed foci, while an empty virus failed to do so. The entire coding region of our ID #7 cDNA was sequenced, revealing no point mutations or deletions compared to the published IHH cDNA sequence. Although activation of Hedgehog (Hh) pathways has been revealed among a wide range of digestive tract cancers,( 13 ) oncogenic activity of IHH has not been reported to date. We supposed from our data that overexpression of IHH may contribute directly to malignant transformation.
Figure 1.
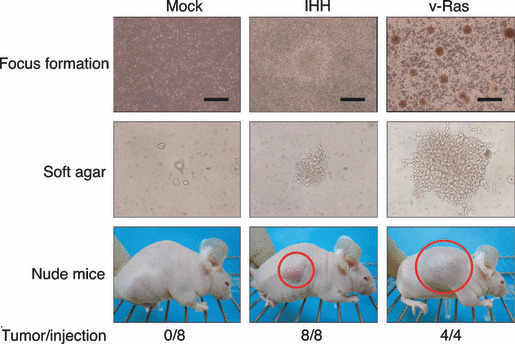
Transforming activity of Indian hedgehog homolog (IHH). Mouse 3T3 cells were infected with viruses encoding IHH or v‐Ras or with the empty virus (Mock), and were then cultured for 5 days for the analysis of focus formation (top panels; scale bars = 1 mm). The same batches of 3T3 cells were also assayed for anchorage‐independent growth in soft agar over 17 days (middle panels) and for tumorigenicity in nude mice over 3 weeks (bottom panels). Tumors formed in the shoulders of mice injected subcutaneously with 1 × 105 cells are indicated by red circles. The frequency of tumor formation (tumors/injection) is also indicated.
Confirmation of the transforming activity of IHH. To confirm the oncogenic activity of IHH, we examined its effect on the anchorage‐independent growth of 3T3 cells in soft agar. Whereas cells infected with an empty virus did not grow in the agar, those infected with a virus expressing IHH formed multiple foci in repeated experiments (Fig. 1, middle panel). In addition, 3T3 cells expressing v‐Ras readily grew in the agar.
The transforming activity of IHH was also tested by the tumor formation assay with athymic nude mice. 3T3 cells infected with the empty virus or retrovirus expressing IHH or v‐Ras were inoculated subcutaneously into nude mice. As shown in the bottom panel of Fig. 1, tumor formation was readily observed for the cells expressing IHH or v‐Ras. These results clearly revealed an unexpected, direct transforming potential of IHH in fibroblasts.
Overexpression of IHH mRNA. Given the transforming potential of wild‐type IHH (when it is abundantly expressed), we tried to examine if IHH is overexpressed in BTC specimens. Real‐time RT‐PCR analysis for the quantitation of IHH cDNA among normal gall bladder (n = 7) and BTC specimens (n = 6) (Supporting Information Table S1) revealed that IHH is indeed overexpressed in the latter specimens, albeit with marginal statistical significance (P = 0.06) by a two‐tailed t‐test (Fig. 2). It should be noted, however, that BTC cases with APBDJ (n = 2) had significantly abundant expression of IHH compared to BTC without APBDJ (P = 0.005) or to normal gall bladder (P = 2.4 × 10−6). Therefore, it is likely that some types of BTC overexpress IHH.
Figure 2.
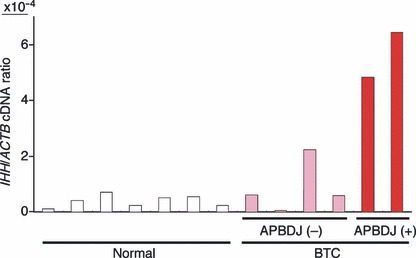
Expression of Indian hedgehog homolog (IHH) in biliary tract. Oligo(dT)‐primed cDNA was synthesized from clinical specimens of biliary tract cancer (BTC) with (+) or without (−) anomalous pancreaticobiliary duct junction (APBDJ), or from normal gallbladder (Normal), and were subjected to quantitative PCR analysis for cDNA of IHH and β‐actin (ACTB). The relative expression level of the former to the latter is represented.
Protein expression of IHH. To confirm the elevated expression of IHH in BTC, we examined its protein level by an immunohistochemical approach. In accordance with the RT‐PCR experiments, IHH protein was abundantly detected only in the cytoplasm of cancerous duct but not in stromal cells for BTC with APBDJ (Fig. 3). We failed to observe such staining in normal gallbladder, suggesting that IHH protein was markedly induced in BTC with APBDJ compared to normal gallbladder.
Figure 3.
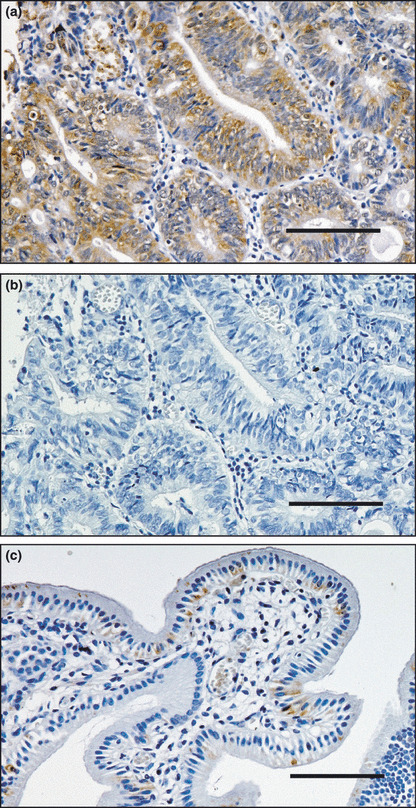
Immunohistochemical detection of Indian hedgehog homolog (IHH). Expression of IHH is elevated in (a) biliary tract cancer with anomalous pancreaticobiliary duct junction, but such reactivity was absent in the control experiment for (b) the same specimen or (c) anti‐IHH staining in normal gallbladder. Scale bars = 100 μm.
Discussion
In the present study, we have constructed a retroviral cDNA expression library for a BTC specimen with APBDJ, and unexpectedly revealed the transforming potential of IHH through a focus formation assay with the mouse fibroblast cell line 3T3. As there were no sequence alterations in our isolated IHH cDNA, the high expression of IHH is likely to exert its oncogenic activity. Consistent with this notion, expression of IHH was indeed activated in BTC with APBDJ.
In our transformation assays for IHH (i.e. focus formation assay, soft agar‐growth assay, and nude mouse‐tumorigenicity assay) we directly used a highly polyclonal, mass culture of 3T3 cells infected with a retrovirus expressing IHH, without any selection (such as positive selection for neomycin resistance‐cells). Repeated confirmation of the transforming potential for IHH in such assays (and not for an empty virus) strongly argues against a hypothesis that an artificial expression of mouse genes adjacent to the retroviral integration sites was responsible for the 3T3 transformation in these experiments.
The Hh signaling pathway was originally described in the development of Drosophila melanogaster as a segment polarity gene required for embryonic patterning.( 14 ) There are three vertebrate homologues of Hh: Ihh, Sonic hedgehog (Shh), and Desert hedgehog (Dhh) with similar biological properties among them. Hh signaling is known to play a pivotal role in cell fate decisions,( 15 ) tissue repair,( 16 ) and stem cell self renewal.( 17 , 18 ) Aberration in such signaling may contribute to sustained cell growth and cancer. Indeed, Hahn et al. and Johnson et al. revealed that mutations within PTCH1 (a binding partner of hedgehog) cause a cancer‐promoting condition, Gorlin syndrome.( 19 , 20 ) Further, frequent mutations in Hh signaling components have also been identified among sporadic basal cell carcinoma( 21 ) and medulloblastoma.( 22 )
In addition, transcriptional activation of Hh components has been demonstrated among a wide range of gastrointestinal tumors, which results from endogenous overexpression of Hh proteins such as IHH and SHH.( 13 ) Despite the lack of gene mutations for the Hh components in these tumors, cyclopamine, a specific inhibitor for SMO, suppresses the growth of tumors positive for elevated Hh signaling, supporting the idea that overexpression of the Hh family of proteins may have a mitogenic function.
Our current data proves for the first time the direct transforming potential of IHH, at least in fibroblasts. Furthermore, apparent overexpression of IHH in BTC with APBDJ indicates an important role of IHH especially in this subtype of BTC. In addition to the presence or absence of APBDJ, we also examined the clinicopathological features of the BTC specimens used in our study. As shown in Supporting Information Table S1, none of the TNM stage, clinical stage, KRAS mutation, or Ki‐67 index were related to the overexpression of IHH. However, because the current cohort size is still small, a larger cohort study is mandatory to examine the clinical features of BTC with high IHH.
Although Yang et al. reported that treatment with a SMO inhibitor leads to downregulation of CCND1 and upregulation of CDKN1A in a cell line of pancreatic carcinoma,( 23 ) we did not observe such a relationship between CCND1/CDKN1A and IHH expression (data not shown). However, overexpression of CCND1 may be more prevalent among BTC than that of IHH,( 24 ) suggesting the presence of an IHH‐independent regulatory network for CCND1 in BTC.
APBDJ causes pancreatic fluid regurgitation into the biliary duct, and is found frequently among BTC cases.( 25 ) Because pancreatic fluid is rich in various proteases, frequent regurgitation of such fluid into the biliary tract is likely to cause sustained inflammation in the tract. Because inflammation and tissue repair cause transcriptional activation of the Hh family of soluble factors,( 16 ) it may not be surprising to find an elevated level of IHH mRNA in the biliary tract with APBDJ. Given the transforming function of abundant IHH, such overexpression may lead to increased cell cycle of biliary tract cells, and eventually to the generation of BTC. Because a number of chemical inhibitors are under development for the Hh pathways,( 26 ) BTC with APBDJ would be an intriguing candidate for such drugs. Further, it is also tempting to examine the Hh ligand levels among human cancers associated with chronic inflammation or regeneration.
Abbreviations
- CCND1
cyclin D1
- CDKN1A
cyclin‐dependent kinase inhibitor 1A
- ERBB2
v‐erb‐b2 avian erythroblastic leukemia viral oncogene homolog 2
- KRAS
v‐ki‐ras2 Kirsten rat sarcoma viral oncogene homolog
- PTCH1
Patched, Drosophila, homolog of, 1
- TP53
tumor protein p53
Supporting information
Table S1. Clinical characteristics of the patients with biliary tract cancer (BTC).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
This work was supported in part by grants for Research on Human Genome and Tissue Engineering and for Third‐Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare of Japan, as well as by a grant for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1. National Cancer Center . Cancer statistics in Japan 2007 (Website on the internet). Tokyo, Japan: National Cancer Center, 2008. [Cited 16 November 2007.] Available from URL: http://www.ganjoho.jp/public/statistics/backnumber/2007_en.html. [Google Scholar]
- 2. Carriaga MT, Henson DE. Liver, gallbladder, extrahepatic bile ducts, and pancreas. Cancer 1995; 75: 171–90. [DOI] [PubMed] [Google Scholar]
- 3. Cubertafond P, Gainant A, Cucchiaro G. Surgical treatment of 724 carcinomas of the gallbladder. Results of the French Surgical Association Survey. Ann Surg 1994; 219: 275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shaib Y, El‐Serag HB. The epidemiology of cholangiocarcinoma. Semin Liver Dis 2004; 24: 115–25. [DOI] [PubMed] [Google Scholar]
- 5. Zatonski WA, Lowenfels AB, Boyle P et al. Epidemiologic aspects of gallbladder cancer: a case‐control study of the SEARCH Program of the International Agency for Research on Cancer. J Natl Cancer Inst 1997; 89: 1132–8. [DOI] [PubMed] [Google Scholar]
- 6. Hasumi A, Matsui H, Sugioka A et al. Precancerous conditions of biliary tract cancer in patients with pancreaticobiliary maljunction: reappraisal of nationwide survey in Japan. J Hepatobiliary Pancreat Surg 2000; 7: 551–5. [DOI] [PubMed] [Google Scholar]
- 7. Rosen CB, Nagorney DM, Wiesner RH, Coffey RJ Jr, LaRusso NF. Cholangiocarcinoma complicating primary sclerosing cholangitis. Ann Surg 1991; 213: 21–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aaronson SA. Growth factors and cancer. Science 1991; 254: 1146–53. [DOI] [PubMed] [Google Scholar]
- 9. Soda M, Choi YL, Enomoto M et al. Identification of the transforming EML4–ALK fusion gene in non‐small‐cell lung cancer. Nature 2007; 448: 561–6. [DOI] [PubMed] [Google Scholar]
- 10. Hatanaka H, Takada S, Choi YL et al. Transforming activity of purinergic receptor P2Y, G‐protein coupled, 2 revealed by retroviral expression screening. Biochem Biophys Res Commun 2007; 356: 723–6. [DOI] [PubMed] [Google Scholar]
- 11. Fujiwara S, Yamashita Y, Choi YL et al. Transforming activity of purinergic receptor P2Y, G protein coupled, 8 revealed by retroviral expression screening. Leuk Lymphoma 2007; 48: 978–86. [DOI] [PubMed] [Google Scholar]
- 12. Choi YL, Kaneda R, Wada T et al. Identification of a constitutively active mutant of JAK3 by retroviral expression screening. Leuk Res 2007; 31: 203–9. [DOI] [PubMed] [Google Scholar]
- 13. Berman DM, Karhadkar SS, Maitra A et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature 2003; 425: 846–51. [DOI] [PubMed] [Google Scholar]
- 14. Nusslein‐Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature 1980; 287: 795–801. [DOI] [PubMed] [Google Scholar]
- 15. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–87. [DOI] [PubMed] [Google Scholar]
- 16. Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature 2004; 432: 324–31. [DOI] [PubMed] [Google Scholar]
- 17. Liu S, Dontu G, Wicha MS. Mammary stem cells, self‐renewal pathways, and carcinogenesis. Breast Cancer Res 2005; 7: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S, Dontu G, Mantle ID et al. Hedgehog signaling and Bmi‐1 regulate self‐renewal of normal and malignant human mammary stem cells. Cancer Res 2006; 66: 6063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hahn H, Wicking C, Zaphiropoulous PG et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 1996; 85: 841–51. [DOI] [PubMed] [Google Scholar]
- 20. Johnson RL, Rothman AL, Xie J et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 1996; 272: 1668–71. [DOI] [PubMed] [Google Scholar]
- 21. Reifenberger J, Wolter M, Weber RG et al. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res 1998; 58: 1798–803. [PubMed] [Google Scholar]
- 22. Taylor MD, Liu L, Raffel C et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet 2002; 31: 306–10. [DOI] [PubMed] [Google Scholar]
- 23. Yang Y, Tian X, Xie X, Zhuang Y, Wu W, Wang W. Expression and regulation of hedgehog signaling pathway in pancreatic cancer. Langenbecks Arch Surg 2009; doi: DOI: 10.1007/s00423-009-0493-9. [DOI] [PubMed] [Google Scholar]
- 24. Hui AM, Li X, Shi YZ, Takayama T, Torzilli G, Makuuchi M. Cyclin D1 overexpression is a critical event in gallbladder carcinogenesis and independently predicts decreased survival for patients with gallbladder carcinoma. Clin Cancer Res 2000; 6: 4272–7. [PubMed] [Google Scholar]
- 25. Kimura K, Ohto M, Saisho H et al. Association of gallbladder carcinoma and anomalous pancreaticobiliary ductal union. Gastroenterology 1985; 89: 1258–65. [DOI] [PubMed] [Google Scholar]
- 26. Rubin LL, De Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 2006; 5: 1026–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinical characteristics of the patients with biliary tract cancer (BTC).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


