Figure 1.
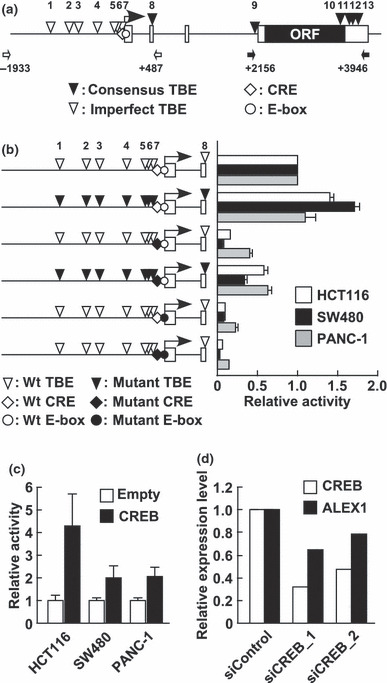
The cyclic AMP response element (CRE) site and CRE‐binding protein (CREB) are involved in human Arm protein lost in epithelial cancers, on chromosome X 1 (ALEX1) regulation. (a) A schematic representation of the genomic structure of the ALEX1 gene on human chromosome X. Open and filled boxes represent the exons of the ALEX1 gene and the ORF encoding the ALEX1 protein, respectively. The bent arrow indicates the transcription start site of the ALEX1 gene. The positions of consensus (CTTTGA/TA/T and A/TA/TCAAAG) and imperfect TCF/LEF‐binding element (TBE)s are indicated by filled and open reverse triangles, respectively, and numbered. An open diamond and circle represent the putative CRE (TGACGTG) and E‐box (CACGTG) site, respectively. The open and closed arrows indicate the primers used for promoter and intron 3‐exon 4 regions of the ALEX1 gene, respectively. (b) In the left diagram, open reverse triangles represent wild‐type TBE sites, and filled reverse triangles represent mutated TBE sites. Open and filled diamonds represent the wild‐type and mutant type of CRE, respectively. Open and filled circles represent the wild‐type and mutant type of E‐box, respectively. The right bar graph represents the relative luciferase activities in HCT116, SW480, and PANC‐1 cells transfected with the reporter plasmid driven by wild‐type and a series of site‐directed mutant type of the ALEX1 promoter. Error bars indicate the SD. (c) Luciferase reporter assay of HCT116, SW480, and PANC‐1 cells co‐transfected with wild‐type ALEX1 promoter‐driven reporter plasmid and the expression plasmid of the indicated genes. Error bars indicate the SD. (d) Quantitative real‐time RT‐PCR analysis of endogenous CREB and ALEX1 mRNA expression in PANC‐1 cells transfected with control siRNA and siRNAs targeting CREB. Relative expression level of CREB and ALEX1 mRNA for each sample was normalized against the expression level of GAPDH mRNA. The average of two independent experiments is shown.
