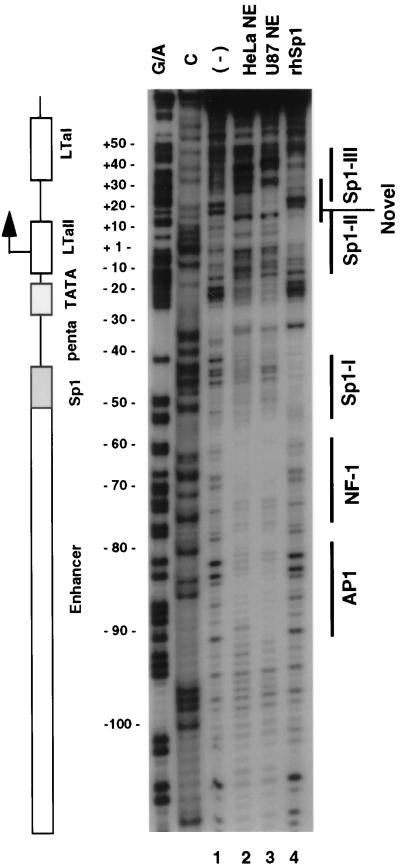
FIG. 4.
DNase I footprinting analysis of the proximal promoter region of JC virus. Nuclear extracts from U87MG and HeLa cells and recombinant human Sp1 were used for DNase I footprinting. The coding strand probe was labeled as described in Materials and Methods. The labeled probe was digested with DNase I in the absence (lane 1) or presence of nuclear extracts prepared from U87MG (lane 2) or HeLa (lane 3) cells or recombinant human Sp1 protein (rhSp1; lane 4). A schematic diagram corresponding to the digested region of the promoter is also presented. Areas of protection were identified by both nuclear extracts in the enhancer region, corresponding to AP1 and NF-1 sites. Areas of protection were noted in the basal promoter region, most of which showed a cell-specific pattern. Three Sp1 binding domains were also identified, as marked by solid lines (Sp1-I, Sp1-II, Sp1-III) at the right.