Abstract
To evaluate the effect of galectin‐3 in cell cycle regulation of colon cancer cells, we looked for binding molecules interacting with galectin‐3 and examined the changes in cell cycle by suppressing galectin‐3 and the binding molecule. To identify target molecules interacting with galectin‐3, we analyzed immunoprecipitate of the anti‐galectin‐3 antibody obtained from human colon cancer cell line, using matrix‐assisted laser desorption ionization‐mass spectrometry. We validated subcellular localization of galectin‐3 and ATP synthase identified, and ATP synthase activity was determined in the presence of galectin‐3. Cell cycle regulation was monitored after galectin‐3 siRNA transfection. ATP synthase b‐subunit was identified in immunoprecipitate of the anti‐galectin‐3 antibody. Galectin‐3 and ATP synthase were co‐isolated in the inner membrane vesicles of mitochondria. Galectin‐3 has an inhibitory activity against ATP synthase, and intracellular ATP content showed increasing tendency after galectin‐3 suppression. Suppression of galectin‐3 resulted in G0/G1 progression of human colon cancer cells arrested at S, S/G2 and G2/M phase in the presence of doxorubicin, and etoposide or nocodazole, respectively. Compared to cells in which ATP synthase d‐subunit was suppressed alone, sub‐G1 fraction caused by etoposide or nocodazole was decreased in cells with galectin‐3 suppression alone. In conclusion, galectin‐3 co‐localized with ATP synthase in the inner membrane of mitochondria and has an inhibitory effect on ATP synthase in human colon cancer cells. In the presence of cell cycle synchronizing drugs, doxorubicin, etoposide, or nocodazole, suppression of galectin‐3 induced cell cycle progression to G0/G1 phase. (Cancer Sci 2008; 99: 1884–1891)
Galectin‐3, previously described as IgE binding protein, CBP35, CBP30, Mac‐2, L‐29, L‐31, and L‐34, is one of the β‐galactoside‐binding proteins that bind to the carbohydrate portion of cell‐surface glycoproteins and glycolipids.( 1 ) In adults, galectin‐3 is expressed ubiquitously in many normal tissues, and is mainly related to the epithelial cells and myeloid cells, including small intestinal epithelial cells, colonic epithelia, and epithelial cells of the kidney, lung, breast, and prostate.( 2 ) Galectin‐3 is found intracellularly in the nucleus and cytoplasm, on the cell surface, and in the extracellular space.( 2 , 3 ) Through specific interactions with many proteins, galectin‐3 is involved in numerous biological processes, such as cell adhesion, proliferation, differentiation, angiogenesis, and apoptosis.( 2 )
A number of studies have reported the roles of galectin‐3 in many types of cancer. In cancer, galectin‐3 has been reported to correlate with malignant transformation, regulation of apoptosis, immune escape, angiogenesis, tumor invasiveness, and metastasis.( 4 , 5 , 6 , 7 , 8 ) The anti‐ or pro‐apoptotic effect is the most extensively studied function of galectin‐3 relevant to tumor progression.( 9 , 10 ) Intracellular galectin‐3 shows activity to inhibit anoikis (apoptosis induced by the loss of cell anchorage) and apoptosis induced by chemotherapeutic agents such as cisplatin, genistein, tumor necrosis factor, and nitric oxide.( 11 ) Interestingly, the activities of galectin‐3 seem different according to subcellular location, showing that nuclear localization of galectin‐3 is mainly related to pro‐apoptotic activity and antitumor effects, whereas its cellular localization away from the nucleus correlates with antiapotosis, tumor progression, and neoplastic progression.( 12 , 13 , 14 ) These findings were characterized based on cancer cells other than colorectal cancer, such as breast, thyroid, and prostate cancers.( 4 , 5 , 6 , 7 , 8 ) In colon cancer, published data regarding galectin‐3 is not yet clear. Some studies have reported a correlation between tumor progression and an increased level of galectin‐3, whereas others have shown opposite results in colon cancer cells.( 15 , 16 , 17 , 18 , 19 ) The function of galectin‐3 is still controversial regarding apoptosis and cell‐cycle regulation in colon cancer cells.
To evaluate the effect of galectin‐3 on the cell‐cycle regulation of colon cancer cells, we looked for binding molecules that interact with galectin‐3 and examined changes in the cell cycle caused by suppressing galectin‐3 and the binding molecule in colon cancer cell lines.
Materials and Methods
Human colon cancer cell lines. The human colon cancer cell lines SNU‐81, SNU‐C4, SNU‐C5, SNU‐769A, SNU‐769B, SNU‐1033, LoVo, and DLD‐1 were obtained from the Korean Cell Line Bank (Seoul, Korea). SNU‐769B was used for immunoprecipitation, matrix‐assisted laser desorption ionization–mass spectrometry (MALDI‐MS), western blot analysis, and a mitochondrial F1F0‐ATP synthase (ATP synthase) activity assay. SNU‐81, SNU‐C4, SNU‐C5, SNU‐769A, SNU‐1033, LoVo, and DLD‐1 were used for western blot analysis. SNU‐81 was also used for small interfering RNA (siRNA) transfection as it shows attached cell growth as well as higher ATP synthase and galectin‐3 expression.( 20 )
Immunoprecipitation. All procedures were carried out at 4°C, unless otherwise specified. Approximately 107 cells in 1 mL cold 1× RIPA containing protease inhibitors (Roche Diagnostics, Mannheim, Germany) were placed on ice for 30 min with occasional mixing. The cell lysate was centrifuged at 13400 g for 10 min, and the supernatant was collected carefully without disturbing the pellet. The supernatant was mixed with primary antibody against either galectin‐3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), ATP synthase α‐subunit (Molecular Probes, Eugene, OR, USA), or d‐subunit (Molecular Probes), and incubated for 2 h on a rocking platform. Prepared protein G sepharose beads (100 µL; GE Health Care Life Sciences, Uppsala, Sweden) were added and further incubated on ice for 1 h on a rocking platform. The mixture was centrifuged at 13400 g for 30 s, and the supernatant carefully removed. Protein G sepharose beads were washed five times with 1 mL cold 1× RIPA to minimize background. Following the last wash, 100 µL 2× sodium dodecylsulfate (SDS) sample buffer was added to the bead pellet, which was heated to 100°C for 10 min. After boiling, the immunoprecipitate was centrifuged at 13400g for 5 min and the supernatant collected for western blot analysis.
Matrix‐assisted laser desorption ionization–mass spectrometry. Sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) gels containing proteins of interest were excised, destained, with 50% acetonitrile in 0.1 mol/L ammonium bicarbonate, and dried in a Speedvac evaporator (Savant Speedvac Plus SC210A Savant Instruments Inc., Holbrook, NY, US). Dried gel pieces were rehydrated with 30 µL of 25 mmol/L sodium bicarbonate, pH 8.8, containing 50 ng trypsin (Promega, Madison, WI, USA) at 37°C overnight. α‐Cyano 4 hydroxycinnamic acid (20 mg; Bruker Daltonics, Bremen, Germany) was dissolved in 1 mL acetone:ethanol (1:2 v/v), and 0.5 µL matrix solution was mixed with an equivalent volume of sample. The sample was analyzed using an Ultraflex TOF–TOF (time of flight) system (Bruker Daltonics). The Ultraflex TOF–TOF system was operated in positive ion reflect mode. Each spectrum was the cumulative average of 250–450 laser shots. Mass spectra were first calibrated in the closed external mode using the peptide calibration standard II (Bruker Daltonics), sometimes using the internal statistical mode to achieve maximum calibration mass accuracy, and analyzed with FlexAnalysis software, version 2.4 (Bruker Daltonics). Peptide mass peaks from each spectrum were submitted to the Mascot peptide mass fingerprinting search form (http://www.matrixscience.com) for analysis with BioTools software, version 3.0 (Bruker Daltonics).
The search included peaks with a signal‐to‐noise ratio greater than 3. The peak list for each sample was sent in and used to query the non‐redundant Mass Spectrometry Protein Sequence Database for protein identification. Standard settings included the following: enzyme, trypsin; missed cleavage, one; fixed modifications, none selected; variable modifications, oxidized methionine; protein mass, blank; mass values, MH + (monoisotopic protonated molecule); mass tolerance, varied between 75 and 100 p.p.m.
Western blot analysis. Western blot analysis was carried out as described earlier.( 20 ) Briefly, 4000×g supernatant fractions of cell homogenates containing equivalent amounts of protein were subjected to SDS‐PAGE. Following electrophoresis, proteins were transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA), which were blocked by incubation for 2 h at 4°C in 1% Tween 20‐TBS (Tris‐buffered saline) containing 1.5% non‐fat dry milk (Bio‐Rad, Richmond, CA, USA) and 1 mmol/L MgCl2. Membranes were incubated for 2 h at room temperature with primary antibodies against either galectin‐3 (1:2000) (Santa Cruz Biotechnology), F1F0‐ATP synthase (ATP synthase) α‐subunit (1:2000) (Molecular Probes), d‐subunit (1:1000) (Molecular Probes), IF1 (1:2000) (Molecular Probes), or actin (1:2000) (Sigma‐Aldrich, St Louis, MO, USA). Membranes were washed three times for 15 min each with blocking solution, and incubated with diluted horseradish peroxidase (HRP)‐conjugated secondary antibody (Southern Biotech, Birmingham, AL, USA) for 1 h at room temperature. Membranes were rewashed (3 × 15 min) with blocking solution, incubated with WEST‐ZOL plus chemiluminescence reagent (Intron Biotechnology, Gyeonggi, Korea) for 1 min, and exposed to film (Kodak Blue XB‐1; Kodak, Rochester, NY, USA).
Isolation of ATP synthase. For isolation of an enriched, functional mitochondrial fraction from the SNU‐769B cell line, a mitochondria isolation kit (Sigma‐Aldrich) was used as recommended by the manufacturer.
Inner‐membrane vesicles of mitochondria were prepared as described previously.( 21 ) Isolated mitochondria were suspended in distilled water and centrifuged at 10 000g for 15 min. The pellet was suspended in distilled water and subjected to sonication for 2 min at 15‐s intervals on ice. Centrifugation was carried out for 10 min at 10 000g at 4°C. The supernatant was centrifuged at 100 000g for 1 h. The final pellet comprised purified inner membranes essentially free of outer membrane, intracristal space, and matrix activities.
Submitochondrial particles were prepared as described in a previous report.( 22 ) Freshly prepared inner‐membrane vesicles were suspended in buffer containing 220 mmol/L d‐mannitol, 70 mmol/L sucrose, 2 mmol/L HEPES, and 0.5 mg/mL defatted bovine serum albumin, pH 7.4. Inner‐membrane vesicles were diluted to 2 mg/mL in buffer containing 0.15 mol/L K2HPO4, 1 mmol/L ATP, 25 mmol/L ethylenediaminetetraacetic acid (EDTA), 0.5 mmol/L dithiothreitol (DTT), and 5% ethylene glycol, pH 7.9, and centrifuged for 1 h at 225 000g at 4°C. The pellet was suspended in dilution buffer and recentrifuged twice as above. The resulting pellet, comprising submitochondrial particles, was suspended in buffer containing 50 mmol/L Tris, 1 mmol/L ATP, 25 mmol/L EDTA, 0.5 mmol/L DTT, and 5% ethylene glycol, pH 7.9.
Synthesis of IF1 and IF1A core peptides. IF1 (NH2‐LAALKKHHEEEIVHHKK‐COOH) and IF1A (NH2‐LAALKKAAEEEIVHHKK‐COOH) were synthesized by Peptron (Daejeon, Korea).( 23 )
ATP synthase activity assay. Mitochondrial fractions were utilized for determining ATP synthase activity, which was measured in the direction of ATP hydrolysis (ATPase activity) using the continuous spectrophotometric assay,( 20 ) except that 2 mmol/L ethylene glycol‐bis(β‐aminoethyl ether)‐N,N,N′,N′‐tetraacetic acid (EGTA) (Sigma‐Aldrich) replaced EDTA in the reaction mixture. Mitochondrial fractions (enzyme source) were added to reaction mixtures containing 60 mmol/L sucrose, 50 mmol/L Tris, pH 8.0, 50 mmol/L KCl, 4 mmol/L MgCl2, 2 mmol/L ATP, 2 mmol/L EGTA, 1 mmol/L KCN with 0.1 mmol/L NADH, 5 U/mL pyruvate kinase, and 5 U/mL lactate dehydrogenase (Sigma‐Aldrich). The total volume in the cuvette was 1 mL. The blank cuvette contained air. The linear reaction was observed at 340 nm for 2 min at 25°C. One unit of activity was defined as that required for the oxidation of 1 µmol NADH/min/mg at 25°C and pH 7.4 under the above conditions. Recombinant human galectin‐3 protein was purchased from Bioclon (San Diego, CA, USA).
Small interfering RNA synthesis and transfection. The following target sequences were used to generate siRNA (Qiagen, Chatsworth, CA, USA): 5′‐CACGGTGAAGCCCAATGCAAA‐3′ (NM_002306) for galectin‐3; 5′‐TAATAATTATACAGTTAAA‐3′ (NM_006356) for ATP synthase d‐subunit; and 5′‐AATTCTCCGAACGTGTCACGT‐3′ for non‐silencing control. Transfection of siRNA was carried out using HiferFect transfection reagent (Qiagen), according to the manufacturer's protocol. Briefly, 2 µL of 20 µmol/L siRNA solution and 20 mL transfection reagent were incubated in 100 mL serum‐free RPMI‐1640 medium for 10 min to facilitate complex formation. The resulting mixture (final concentration 5 nmol/L) was added to the human colon cancer cell line SNU‐81 (4 × 105 cells) and grown in a 60‐mm tissue culture dish with 4 mL RPMI‐1640 medium.
Measurement of intracellular ATP content. Intracellular ATP content was measured using the ATP Bioluminescence Assay Kit HS II (Roche Diagnostics) as recommended by the manufacturer.
Cell‐cycle analysis. SNU‐81 cells were transfected with siRNA specific for either galectin‐3 or mitochondrial F1F0‐ATP synthase d‐subunit in the presence of the anticancer drugs doxorubicin (0.5 µmol/L; Sigma‐Aldrich), etoposide (100 µmol/L, Sigma‐Aldrich), nocodazole (166 µmol/L, Sigma‐Aldrich), or 5‐fluorouracil (5‐FU) (10 µmol/L, Sigma‐Aldrich). Changes in cell‐cycle regulation were determined using a FACS Calibur Flow Cytometer (Becton Dickinson, San Jose, CA, USA) and CellQuest software (Becton Dickinson).
Statistical analyses. Within‐group correlations were calculated using the Spearman rank coefficient. Significance was set at P < 0.05.
Results
Identification of mitochondrial ATP synthase b‐subunit in immunoprecipitate of the antigalectin‐3 antibody. Immunoprecipitation with the antigalectin‐3 antibody and whole homogenates of the human colon cancer cell line SNU‐769B was carried out. Proteins in the immunoprecipitate were fractionated using 1 and 2 mol/L NaCl RIPA, and finally extracted with 2× SDS sample buffer. Fractionated proteins were subjected to SDS‐PAGE (Fig. 1a). After staining, protein bands were analyzed by MALDI‐MS. ATP synthase b‐subunit was identified unambiguously in the proteins extracted with SDS sample buffer (Fig. 1a,b). Because the anti‐ATP synthase b‐subunit antibody was not commercially available, we selected two antibodies against the α‐ and d‐subunits of the F1 and F0 complexes, respectively, to verify whether galectin‐3 interacts with ATP synthase. The antigalectin‐3 immunoprecipitate was probed with anti‐ATP synthase α‐ or d‐subunit antibody, and vice versa (Fig. 1c). Immunoreactive signals for both ATP synthase d‐subunit and galectin‐3 were clearly detected in immunoprecipitates of antigalectin‐3 or anti‐ATP synthase d‐subunit antibody. However, no signals for the ATP synthase α‐subunit were detected in immunoprecipitates of the antigalectin‐3 antibody (Fig. 1c). Western blot analysis was carried out to determine the total levels of galectin‐3 in eight different human colon cancer cell lines (Fig. 1d, upper panel). The expression level of galectin‐3 was positively correlated with ATP synthase α‐subunit (P = 0.0046), but the expressional correlation between galectin‐3 and ATP synthase d‐subunit did not reach statistical significance (P = 0.1323) (Fig. 1d, lower panel).
Figure 1.
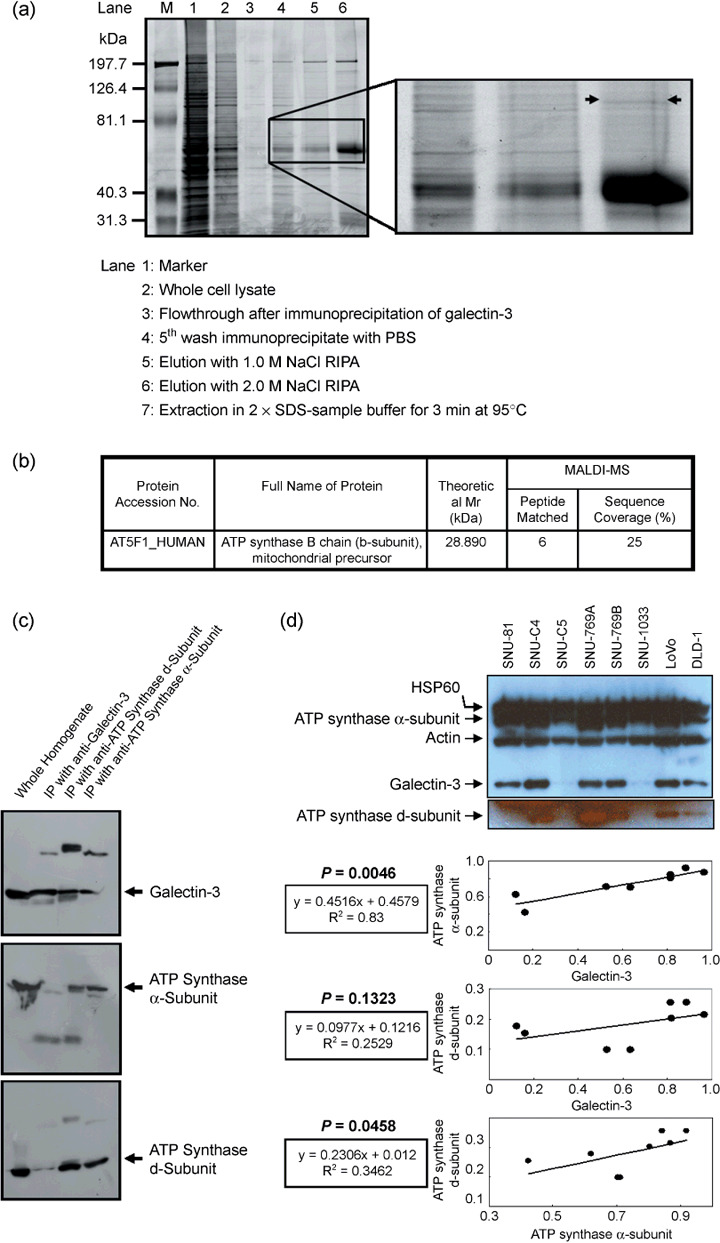
Identification of mitochondrial F1F0‐ATP synthase (ATP synthase) b‐subunit in the antigalectin‐3 antibody immunoprecipitate. (a) Sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) of the galectin‐3 antibody immunoprecipitate. The galectin‐3 immunoprecipitate was fractionated using a salt gradient, as shown in the figure, and subjected to SDS‐PAGE. (b) Identification of the ATP synthase b‐subunit in the immunoprecipitate by matrix‐assisted laser desorption ionization–time of flight analysis. After staining the SDS‐PAGE gel, the protein band specified in (a) was excised and in‐gel digested to determine the peptide mass. The protein was successfully identified as the ATP synthase b‐subunit, a mitochondrial precursor. (c) Interactions of the ATP synthase d‐subunit with galectin‐3. Immunoprecipitates of the antigalectin‐3 antibody were probed with commercially available anti‐ATP synthase α‐ or d‐subunit antibodies, and vice‐versa. Immunoreactive signals of the ATP synthase d‐subunit were clearly detected in immunoprecipitates of antigalectin‐3 and anti‐ATP synthase α‐subunit antibodies. (d) Positive correlation of galectin‐3 expression with ATP synthase α‐ and d‐subunits in eight individual human colon cancer cell lines. Expression of galectin‐3 and ATP synthase subunits was normalized with actin and heat shock protein (HSP) 60, respectively. OD, arbitrary unit.
Coisolation of galectin‐3 with ATP synthase. To determine the presence of galectin‐3 in the mitochondria of human colon cancer cells, the protein level was determined in each isolation step of ATP synthase. The mitochondria were initially isolated from whole homogenates of SNU‐769B, and inner‐membrane vesicles of the mitochondria and submitochondrial particles were prepared, as shown in Figure 2a (upper panel). ATP synthase α‐ and d‐subunits were detected in each fraction. Higher amounts of galectin‐3 were detected in the mitochondrial fraction (Fig. 2a, lower panel). Galectin‐3 was additionally observed in the inner‐membrane vesicles (Fig. 2a, lower panel). In contrast, IF1, an inhibitor protein of ATP synthase, was not detected in the inner‐membrane vesicles of mitochondria (Fig. 2a, lower panel).
Figure 2.
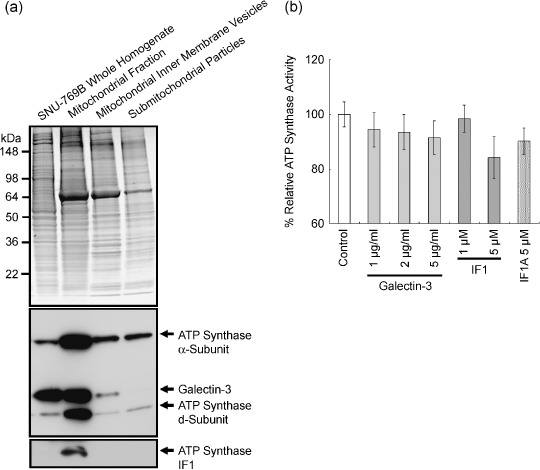
Copurification of galectin‐3 with ATP synthase, and the inhibitory effect of galectin‐3 on ATP synthase activity. (a) Copurification of galectin‐3 with ATP synthase. Mitochondria were isolated initially from whole homogenates of SNU‐769B, and inner‐membrane vesicles of mitochondria and submitochondrial particles were prepared. The upper panel depicts the sodium dodecylsulfate–polyacrylamide gel electrophoresis gel image after staining, and the lower panel represents western blot results. The levels of ATP synthase α‐ and d‐subunits, IF1, and galectin‐3 were determined in each isolation step. Galectin‐3 was additionally detected in the inner‐membrane vesicles (lower panel). (b) Inhibitory effect of galectin‐3 on ATP synthase activity. The control activity of SNU‐769A was defined as 100%, and the percentage relative ATP synthase activities have been determined in the presence of either ATP synthase inhibitor or galectin‐3. Enzyme activity was decreased by up to 8% in the presence of 2 µg/mL galectin‐3.
Interaction of galectin‐3 with ATP synthase, and its inhibitory effect on ATP synthase activity. To clarify the role of galectin‐3 in the mitochondria, ATP synthase activity was measured in its presence. No dramatic change in enzyme activity was evident after adding galectin‐3 to the enzyme reaction mixture. However, enzyme activity was decreased by up to 8% upon the addition of 2 µg/mL galectin‐3 (less than 0.2 µmol/L). This inhibitory effect was almost half of that induced by 5 µmol/L of the IF1 core peptide (Fig. 2b). However, IF1A, the IF1 core peptide with replacement of His48 and His49 with Ala, showed a less‐inhibitory effect on ATP synthase compared to that of the IF1 core peptide (Fig. 2b).
The intracellular ATP content was monitored after suppression of galectin‐3 activity. The human colon cancer cell line SNU‐81 was selected for this experiment as higher transfection efficacy of siRNA was verified in our previous experiments.( 20 ) Following transfection of siRNA specific for galectin‐3, expression of the protein decreased, and was almost completely suppressed at 76 h after transfection (Fig. 3a). Intracellular ATP content was determined at 96 h after siRNA transfection (Fig. 3b). Intracellular ATP in cells transfected with galectin‐3 siRNA was increased compared with the controls transfected with buffer only or non‐silencing siRNA. However, these changes were not statistically significant (Fig. 3b).
Figure 3.
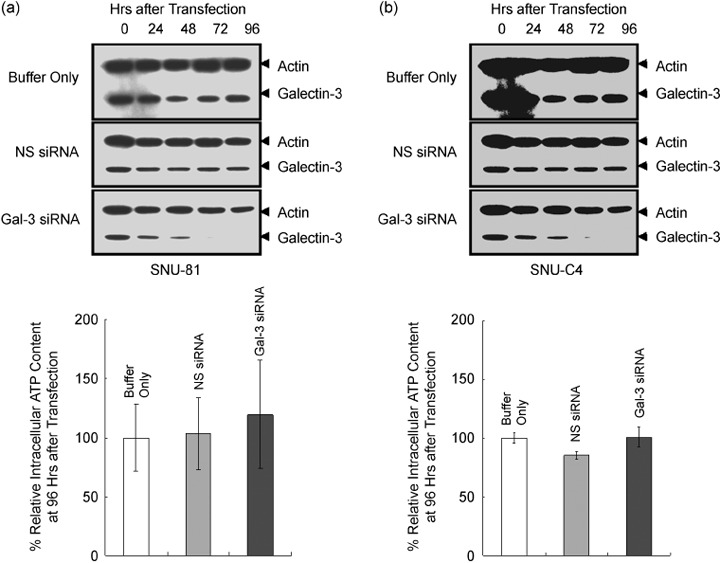
Effects of galectin‐3 suppression on intracellular ATP content of SNU‐81. (a) Suppression of galectin‐3 after small interfering RNA (siRNA) transfection. The human colon cancer cell line SNU‐81 was transfected with siRNA specific for galectin‐3. Galectin‐3 siRNA transfection resulted in suppression of both of galectin‐3 and ATP synthase d‐subunit. (b) Intracellular ATP content after galectin‐3 suppression. The intracellular ATP level in cells transfected with galectin‐3 siRNA increased slightly compared to the controls, but data were not statistically significant.
Effect of ATP synthase and galectin‐3 suppression on cell‐cycle arrest in colon cancer cells. Expression of galectin‐3, ATP synthase d‐subunit, or both proteins were successfully reduced in SNU‐81 by specific siRNA transfection (Fig. 4a). In the absence of anticancer drugs, transfection of siRNA specific to ATP synthase d‐subunit or galectin‐3 or did not affect cell‐cycle progression; however, compared to other test sets an increased G0–G1 population (~10%) was observed with galectin‐3 suppression (Fig. 4b).
Figure 4.
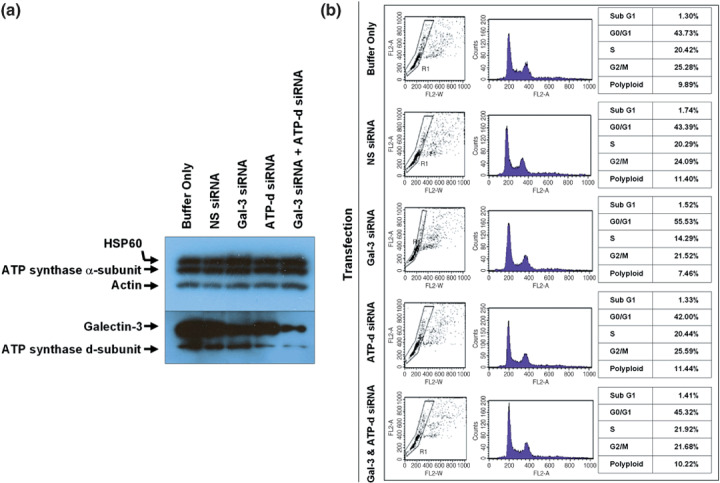
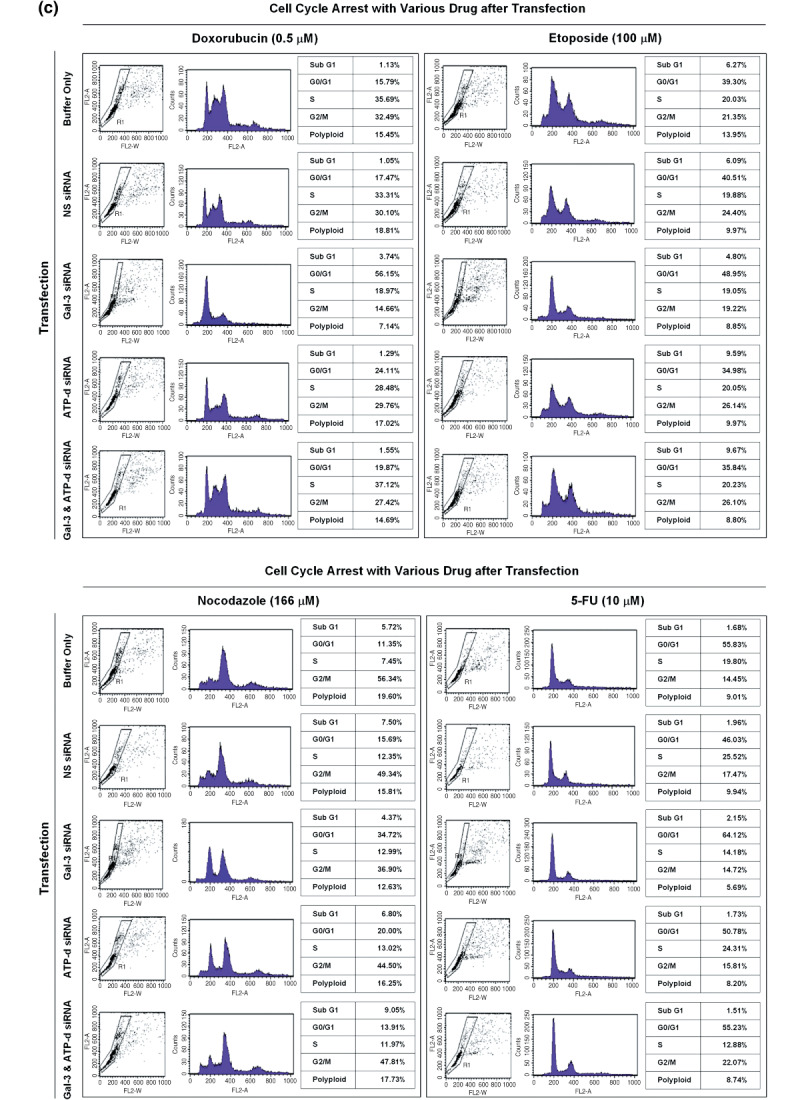
Effect of ATP synthase and galectin‐3 suppression on SNU‐81 cell‐cycle arrest. (a) Suppression of galectin‐3 and ATP synthase d‐subunit after small interfering RNA transfection. (b) There was no significant effect of galectin‐3 or ATP synthase d‐subunit expression on cell‐cycle progression. (c) Cell‐cycle arrest caused by doxorubicin, etoposide, and nocodazole treatment, and the effect of galectin‐3 and ATP synthase d‐subunit suppression.
In the presence of various drugs, a large proportion of SNU‐81 were arrested a certain phase of the cell cycle (Fig. 4c). Treatment with either etoposide (100 µmol/L) or nocodazole (166 µmol/L) increased the sub‐G1 apoptotic portion of cells (Fig. 4c). However, doxorubicin (0.5 µmol/L) and 5‐FU (10 µmol/L) did not cause any significant change in the sub‐G1 population (Fig. 4c). A decreased G0–G1 population and increased arrest in S and G2–M phase was observed with doxorubicin treatment, and nocodazole led to significant arrest of cells in G2–M phase at the given concentration (Fig. 4c). Compared to other drugs tested, 5‐FU did not cause any change in cell‐cycle progression (Fig. 4c).
Cells transfected with galectin‐3 siRNA alone showed significantly higher G0–G1 progression in the presence of the individual drugs tested (Fig. 4c). Furthermore, the sub‐G1 fraction caused by etoposide or nocodazole treatment was decreased after galectin‐3 suppression, but was increased with doxorubicin treatment (Fig. 4c). ATP synthase d‐subunit siRNA transfection also increased G0–G1 progression after treatment with either doxorubicin or nocodazole, and the sub‐G1 portion in the presence of etoposide was increased by ATP synthase d‐subunit suppression (Fig. 4c).
Interestingly, dual suppression of galectin‐3 and ATP synthase d‐subunit counterbalanced each individual suppression effect in some cases. For example, suppression of ATP synthase d‐subunit or galectin‐3 alone resulted in a significant increase in the G0–G1 population in the presence of nocodazole; however, dual suppression did not have any effect on cell‐cycle progression (Fig. 4c). Individual suppression decreased the cell population arrested in S‐phase caused by the treatment of doxorubicin, but suppression of both galectin‐3 and ATP synthase d‐subunit recovered S‐phase arrest (Fig. 4c). Compared to the controls, cells in which galectin‐3 alone was suppressed had a significantly smaller sub‐G1 fraction than those treated with nocodazole; however, this fraction was significantly increased in cells with both galectin‐3 and ATP synthase d‐subunit suppression (Fig. 4c).
Discussion
Galectin‐3 is located predominantly in the cytoplasm, but is also present in the nucleus, cell surface, and extracellular matrix, explaining its multifunctionality.( 24 ) Galectin‐3 is enriched in the mitochondria, where it regulates mitochondrial integrity and prevents mitochondrial damage by blocking the release of cytochrome c from mitochondria and the activation of caspases. These finding suggest that galectin‐3 is a mitochondria‐associated apoptotic regulator in addition to Bcl‐2.( 25 , 26 ) This location of galectin‐3 is regulated by galectin‐3‐interacting proteins, such as synexin, which is crucial for translocation of galectin‐3 to the mitochondrial membranes.( 27 ) However, contrary to the cytoplasmic and nuclear locations, there are few reports regarding galectin‐3‐interacting proteins in the mitochondria. We first identified mitochondrial F1F0‐ATP synthase (ATP synthase) as a protein interacting with galectin‐3 in the mitochondrial environment and localized galectin‐3 to the inner membrane of the mitochondria (1, 2). Moreover, a positive correlation between galectin‐3 and ATP synthase expression in the present study (Fig. 1d), and the presence of galectin‐3 in the mitochondrial inner membrane might support the role of galectin‐3 in maintaining mitochondrial integrity (3, 4).
ATP synthase is a complex of 16 different subunits, with α, β, γ, δ, and ɛ comprising the F1 component, and a, b, c, d, e, f, g, A6L, oligomycin sensitivity‐conferring protein, and coupling factor 6 comprising F0 and stator.( 28 , 29 ) F0 is a membranous domain, whereas F1 is a catalytic domain. The C‐terminal fraction of the b‐subunit plays a critical role in the functional organization of the proton channel of the F0 complex and oligomycin inhibition of proton conduction in the F0 complex.( 30 ) Identification of the ATP synthase b‐subunit in the immunoprecipitate of antigalectin‐3 antibody by MALDI‐MS (Fig. 1a,b), and the positive immunoreactive signal of the ATP synthase d‐subunit indicate that galectin‐3 binds to F0 rather than the F1 complex, supporting the possibility that the protein affects the general function of ATP synthase (Fig. 1c).
It has been shown that active regulation of ATP synthase exists in response to cellular energy demand.( 30 ) On a molecular level, calcium‐binding inhibitor protein and the inhibitor protein IF1 were reported to mediate upregulation and downregulation of ATP synthase.( 30 , 31 ) The IF1 core peptide used in our present study was as effective as the IF1 whole protein in inhibiting ATP synthase activity, but IF1A, the IF1 core peptide with replacement of His48 and His49 with Ala, showed a rather less‐inhibitory effect on ATP synthase compared to that of IF1 (Fig. 2b).( 23 ) Our present study shows that galectin‐3 also interacts with ATP synthase in the mitochondria of human colon cancer cells, having an inhibitory effect on enzyme activity (Fig. 2b). The results shown in 1, 2 highlight some differences in ATP synthase binding and activity between galectin‐3 and IF1. Galectin‐3 interacts specifically with the F0 complex (Fig. 1c), whereas IF1 is located on one side of the lateral stalk of ATP synthase with a binding site that encompasses both α‐ and β‐subunits.( 31 ) Another difference involves the affinities of these molecules to ATP synthase. During the isolation of ATP synthase, galectin‐3 was identified, even in mitochondrial inner‐membrane vesicles (Fig. 2a), and thus the possibility of a higher affinity of galectin‐3 for ATP synthase, compared to IF1, could not be eliminated. Although suppression of galectin‐3 did not lead to significant changes in intracellular ATP content (Fig. 3), its inhibitory effect on ATP synthase was comparable to that of IF1, the most extensively characterized natural protein inhibitor of ATP synthase to date. Our present report describes a certain portion of galectin‐3 being localized to mitochondria, where it acts as a potential inhibitor of ATP synthase. If the subcellular location of galectin‐3 is limited in mitochondria, transfection of siRNA specific for galectin‐3 may result in the removal of one natural ATP synthase inhibitor in mitochondria, and cause an increase in intracellular ATP. However, before our report, we knew of the presence of a natural ATP synthase inhibitor, IF1, in mitochondria. The number of regulatory mechanisms for ATP synthase or ATP generation in cells remains undetermined. Here we have to accept two points: (1) either ATP synthase or intracellular ATP generation is not controlled by a single molecule in a cell; or (2) a large portion of intracellular ATP molecules are produced in the mitochondria, but they are also generated through various metabolic pathways in cells.
After we identified ATP synthase as a molecule that interacts with galectin‐3, we further evaluated the effect of ATP synthase and galectin‐3 suppression on cancer cell‐cycle arrest. It was not possible to upregulate ATP synthase activity directly by transfecting genes corresponding to complete individual subunits because it is a complex of multiple subunits. Among the various functions of galectin‐3, regulation of apoptosis and the cell cycle are the best studied.( 11 , 32 , 33 ) These studies were based uniformly on human breast carcinoma BT549 cells. BT549 breast cancer cells innately undergo apoptosis by the loss of cell anchorage (anoikos), but overexpression of galectin‐3 prevents them from undergoing apoptosis, suggesting that galectin‐3‐mediated inhibition of anoikis may result from its ability to induce cell‐cycle arrest at late G1 through the expression modulation of cyclins and their inhibitors.( 32 ) Apart from cell‐cycle arrest at late G1 in response to loss of cell adhesion, galectin‐3 also influences G2–M arrest of BT549 cells following genistein treatment.( 33 ) In our experimental model using a colon cancer cell line, nocodazole effectively induced G2–M arrest with apoptosis. However, in cells transfected with siRNA specific for galectin‐3, the cell cycle was promoted to G0–G1 and there was a significant decrease in the sub‐G1 portion (Fig. 4c). Similar results were found in cells treated with other cell cycle arrest drugs, such as etoposide (S–G2 arrest) and doxorubicin (S arrest) (Fig. 4c). However, as ATP synthase activity was inhibited in the presence of galectin‐3, dual suppression of galectin‐3 and ATP synthase d‐subunit counterbalanced each individual suppression effect. At present, the way galectin‐3 regulates expression of the cyclins and their inhibitors is not entirely clear, and further studies are needed.
In conclusion, galectin‐3 colocalized with ATP synthase on the inner membrane of mitochondria and has an inhibitory effect on ATP synthase in human colon cancer cells. In the presence of the cell cycle‐synchronizing drugs doxorubicin, etoposide, and nocodazole, suppression of galectin‐3 and ATP synthase induced cell‐cycle progression to G0–G1 phase and reduced apoptotic cell death.
Acknowledgments
This work was supported by a research grant (0710670‐1) from the National Cancer Center, Korea.
References
- 1. Barondes SH, Castronovo V, Cooper DN et al . Galectins: a family of animal β‐galactoside‐binding lectins. Cell 1994; 76: 597–8. [DOI] [PubMed] [Google Scholar]
- 2. Dumic J, Dabelic S, Flogel M. Galectin‐3: an open‐ended story. Biochimica et Biophysica Acta 2006; 1760: 616–35. [DOI] [PubMed] [Google Scholar]
- 3. Ruebel KH, Jin L, Qian X et al . Effects of DNA methylation on galectin‐3 expression in pituitary tumors. Cancer Res 2005; 65: 1136–40. [DOI] [PubMed] [Google Scholar]
- 4. Paz A, Haklai R, Elad‐Sfadia G et al . Galectin‐1 binds oncogenic H‐Ras to mediate Ras membrane anchorage and cell transformation. Oncogene 2001; 20: 7486–93. [DOI] [PubMed] [Google Scholar]
- 5. Takenaka Y, Fukumori T, Yoshii T et al . Nuclear export of phosphorylated galectin‐3 regulates its antiapoptotic activity in response to chemotherapeutic drugs. Mol Cell Biol 2004; 24: 4395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu FT, Patterson RJ, Wang JL. Intracellular functions of galectins. Biochim Biophys Acta 2002; 1572: 263–73. [DOI] [PubMed] [Google Scholar]
- 7. Takenaka Y, Fukumori T, Raz A. Galectin‐3 and metastasis. Glycoconj J 2004; 19: 543–9. [DOI] [PubMed] [Google Scholar]
- 8. Liu FT, Rabinovich G. Galectins as modulators of tumour progression. Nat Rev Cancer 2005; 5: 29–41. [DOI] [PubMed] [Google Scholar]
- 9. Choi JH, Chun KH, Raz A et al . Inhibition of N‐(4‐hydroxyphenyl) retinamide‐induced apoptosis in breast cancer cells by galectin‐3. Cancer Biol Ther 2004; 3: 447–52. [DOI] [PubMed] [Google Scholar]
- 10. Hoyer KK, Pang M, Gui D et al . An anti‐apoptotic role for galectin‐3 in diffuse large B‐cell lymphomas. Am J Pathol 2004; 164: 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakahara S, Oka N, Raz A. On the role of galectin‐3 in cancer apoptosis. Apoptosis 2005; 10: 267–75. [DOI] [PubMed] [Google Scholar]
- 12. Sanjuan X, Fernandez PL, Castells A et al . Differential expression of galectin 3 and galectin 1 in colorectal cancer progression. Gastroenterology 1997; 113: 1906–15. [DOI] [PubMed] [Google Scholar]
- 13. Honjo Y, Inohara H, Akahani S et al . Expression of cytoplasmic galectin‐3 as a prognostic marker in tongue carcinoma. Clin Cancer Res 2000; 6: 4635–40. [PubMed] [Google Scholar]
- 14. Van Den Brule FA, Waltregny D, Liu FT et al . Alteration of the cytoplasmic/nuclear expression pattern of galectin‐3 correlates with prostate carcinoma progression. Int J Cancer 2000; 89: 361–7. [DOI] [PubMed] [Google Scholar]
- 15. Castronovo V, Campo E, Van Den Brule FA et al . Inverse modulation of steady‐state messenger RNA levels of two non‐integrin laminin‐binding proteins in human colon carcinoma. J Natl Cancer Inst 1992; 84: 1161–9. [DOI] [PubMed] [Google Scholar]
- 16. Lotz MM, Andrews CW, Korzelius CA et al . Decreased expression of Mac‐2 (carbohydrate binding protein 35) and loss of its nuclear localization are associated with the neoplastic progression of colon carcinoma. Proc Natl Acad Sci USA 1993; 90: 3466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Irimura T, Matsushita Y, Sutton RC et al . Increased content of an endogenous lactose‐binding lectin in human colorectal carcinoma progressed to metastatic stages. Cancer Res 1991; 51: 387–93. [PubMed] [Google Scholar]
- 18. Schoeppner HL, Raz A, Ho SB et al . Expression of an endogenous galactose‐binding lectin correlates with neoplastic progression in the colon. Cancer 1995; 75: 2818–26. [DOI] [PubMed] [Google Scholar]
- 19. Nagy N, Legendre H, Engels O et al . Refined prognostic evaluation in colon carcinoma using immunohistochemical galectin finger‐printing. Cancer 2003; 97: 1849–58. [DOI] [PubMed] [Google Scholar]
- 20. Shin YK, Yoo BC, Chang HJ et al . Down‐regulation of mitochondrial F1F0‐ATP synthase in human colon cancer cells with induced 5‐fluorouracil resistance. Cancer Res 2005; 15: 3162–70. [DOI] [PubMed] [Google Scholar]
- 21. Pedersen PL, Hullihen J. Adenosine triphosphatase of rat liver mitochondria. Capacity of the homogeneous F1 component of the enzyme to restore ATP synthesis in urea‐treated membranes. J Biol Chem 1978; 253: 2176–83. [PubMed] [Google Scholar]
- 22. McEnery MW, Buhle EL Jr, Aebi U et al . Proton ATPase of rat liver mitochondria. Preparation and visualization of a functional complex using the novel zwitterionic detergent 3‐[(3‐cholamidopropyl) dimethylammonio]‐1‐propanesulfonate. J Biol Chem 1984; 259: 4642–51. [PubMed] [Google Scholar]
- 23. Papa S, Zanotti F, Cocco T et al . Identification of functional domains and critical residues in the adenosinetriphosphatase inhibitor protein of mitochondrial F0F1 ATP synthase. Eur J Biochem 1996; 240: 461–7. [DOI] [PubMed] [Google Scholar]
- 24. Krzeslak A, Lipinska A. Galectin‐3 as a multifunctional protein. Cell Mol Biol Lett 2004; 9: 305–28. [PubMed] [Google Scholar]
- 25. Matarrese P, Tinari N, Semeraro ML et al . Galectin‐3 overexpression protects from cell damage and death by influencing mitochondrial homeostasis. FEBS Lett 2000; 473: 311–15. [DOI] [PubMed] [Google Scholar]
- 26. Moon BK, Lee YJ, Battle P et al . Galectin‐3 protects human breast carcinoma cells against nitric oxide induced apoptosis. Implication of galectin‐3 function during metastasis. Am J Pathol 2001; 159: 1055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu F, Finley RL, Raz A et al . Galectin‐3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. A role for synexin in galectin‐3 translocation. J Biol Chem 2002; 277: 15 819–27. [DOI] [PubMed] [Google Scholar]
- 28. Collinson IR, Runswick MJ, Buchanan SK et al . F0 membrane domain of ATP synthase from bovine heart mitochondria: purification, subunit composition, and reconstitution with F1‐ATPase. Biochemistry 1994; 33: 7971–8. [DOI] [PubMed] [Google Scholar]
- 29. Chang HJ, Lee MR, Hong SH et al . Identification of mitochondrial F0F1‐ATP synthase involved in liver metastasis of colorectal cancer. Cancer Sci 2007; 98: 1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Champagne E, Martinez LO, Collet X et al . Ecto‐F1F0 ATP synthase/F1 ATPase: metabolic and immunological functions. Curr Opin Lipidol 2006; 17: 279–84. [DOI] [PubMed] [Google Scholar]
- 31. Das AM. Regulation of the mitochondrial ATP‐synthase in health and disease. Mol Genet Metab 2003; 79: 71–82. [DOI] [PubMed] [Google Scholar]
- 32. Kim HR, Lin HM, Biliran H et al . Cell cycle arrest and inhibition of anoikis by galectin‐3 in human breast epithelial cells. Cancer Res 1999; 59: 4148–54. [PubMed] [Google Scholar]
- 33. Lin HM, Moon BK, Yu F et al . Galectin‐3 mediates genistein‐induced G2/M arrest and inhibits apoptosis. Carcinogenesis 2000; 21: 1941–5. [DOI] [PubMed] [Google Scholar]


