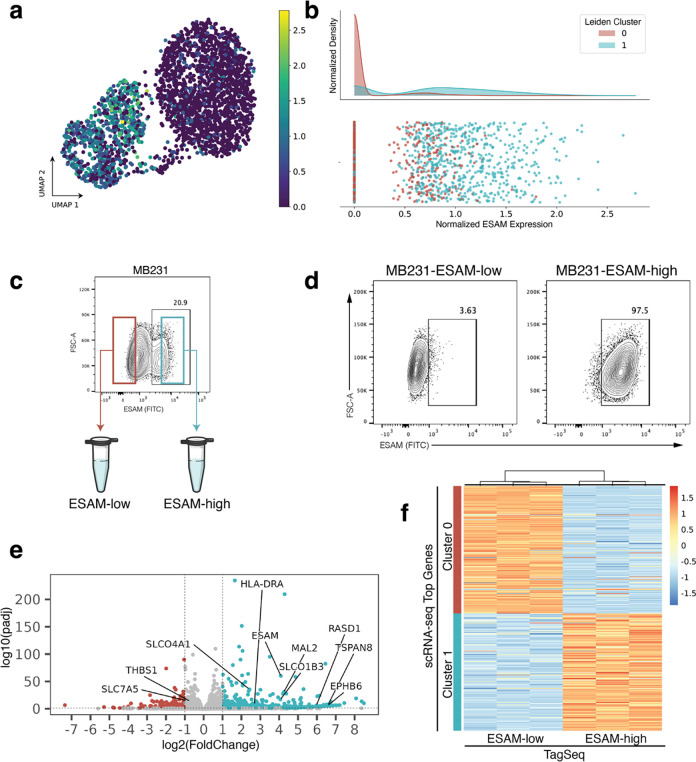
Figure 3. Experimental validation of ESAM as a surface marker for MB231 subpopulations.
(a) scRNA-seq UMAP projection of MB231 cells colored by ESAM expression. (b) Histogram of ESAM expression showing enrichment of cluster 1 for ESAM. (c) ESAM staining on parental MB231 cells. Cells with the lowest staining were FACS enriched as “ESAM-low” and cells with the highest staining as “ESAM-high”. (d) At the time of RNA collection (10 days post-FACS and expansion), sorted ESAM subpopulations each maintained around 97% purity. (e) Volcano plot of bulk Tag-Seq performed on sorted subpopulations. Labels point to genes that were highly ranked in the scRNA-seq dataset using EMD. (f) Heatmap of bulk TaqSeq data for each FACS enriched subpopulation plotted against ranked cluster genes from scRNA-seq.