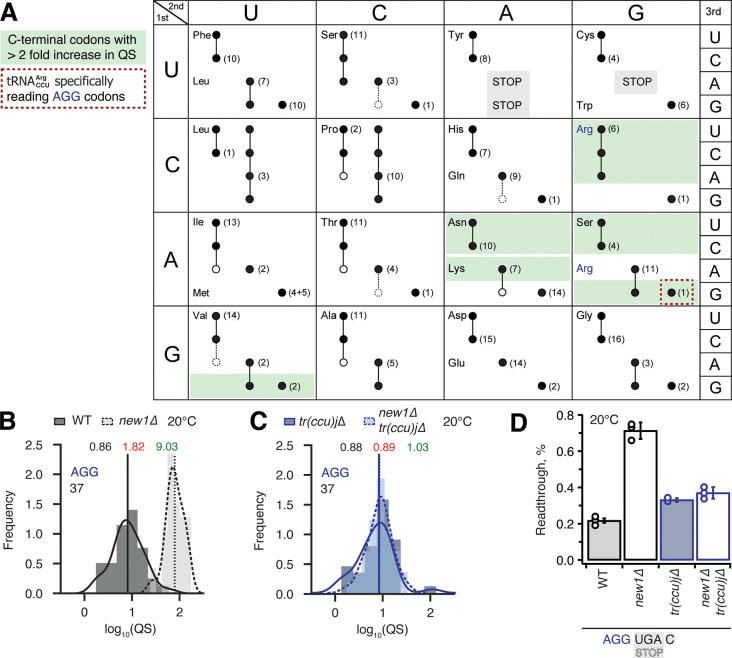
Figure 6. Termination defect in new1Δ yeast is dependent on the nature of the tRNA isoacceptor species present in the P-site of the pre-termination complex.
(A) Decoding abilities of tRNA isoacceptors, adapted from Johansson and colleagues (Johansson et al., 2008). Codons read by individual tRNA species are indicated by circles connected by lines. The gene copy number for each tRNA species is indicated and the position of the number denotes the cognate codon. Empty circles indicate that the tRNA is less likely to read the codon. The empty dashed circles indicate that the tRNA does not efficiently read the codon. C-terminal codons that display increased QS in the new1Δ strain are highlighted with green shading. (B,C) Queuing score distributions for NEW1 (solid outline) and new1Δ (dashed) strains either expressing (B) or lacking (C) . (D) Penultimate AGG codons are not associated with readthrough in new1Δ yeast when decoded by the near-cognate species. Readthrough efficiency for UGA stop codon preceded by penultimate AGG codon in strains either expressing or lacking . All experiments were performed at 20 °C.