Fig. 3. BAP1 deficient pancreatic tumors exhibit hippo pathway deregulation.
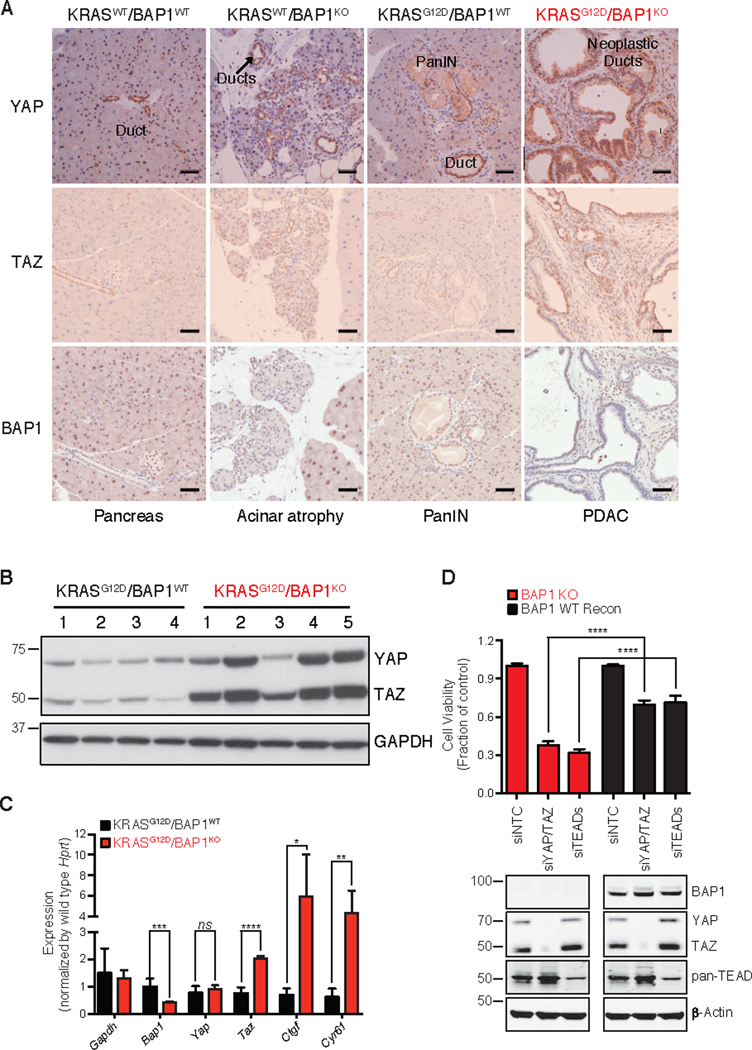
(A) YAP, TAZ, and BAP1 IHC in pancreatic section of 7 week old mice of indicated genotypes. Bar = 50 μM
(B) Immunoblotting analysis of YAP and TAZ from pancreas of 7 week old mice of indicated genotypes. Analysis was performed to detect the indicated proteins. GAPDH served as a loading control.
(C) Levels of Gapdh, Bap1, Yap, Taz and hippo pathway target genes Ctgf and Cyr61 assessed by TaqMan. Hprt was used as a normalization control. Error bars represent mean ± SD (n=3). ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Student’s t test.
(D) Effect of depletion of Yap and Taz in a BAP1 KO pancreatic cancer cell line derived from KRASG12D/BAP1KO tumors. Cells were transiently transfected with siRNAs for Yap, Taz and Teads. Viable cells were determined 6 days later using Cell Titer-Glo and depletion of YAP, TAZ or TEADs was confirmed by immunoblotting. α-Actin served as a loading control. Error bars represent mean ± SD (n=3). ****P < 0.0001; Student’s t test. BAP reconstitution and knockdown of YAP/TAZ or TEADs was confirmed by immunoblotting with indicated antibodies.
