Abstract
Additional copies of chromosome 1 long arm (1q) are frequently found in multiple myeloma (MM) and predict high-risk disease. Available data suggest a different outcome and biology of patients with amplification (Amp1q, ≥4 copies of 1q) vs. gain (Gain1q, 3 copies of 1q) of 1q. We evaluated the impact of Amp1q/Gain1q on the outcome of newly diagnosed MM patients enrolled in the FORTE trial (NCT02203643). Among 400 patients with available 1q data, 52 (13%) had Amp1q and 129 (32%) Gain1q. After a median follow-up of 62 months, median progression-free survival (PFS) was 21.2 months in the Amp1q group, 54.9 months in Gain1q, and not reached (NR) in Normal 1q. PFS was significantly hampered by the presence of Amp1q (HR 3.34 vs. Normal 1q, P < 0.0001; HR 1.99 vs. Gain1q, P = 0.0008). Patients with Gain1q had also a significantly shorter PFS compared with Normal 1q (HR 1.68, P = 0.0031). Concomitant poor prognostic factors or the failure to achieve MRD negativity predicted a median PFS < 12 months in Amp1q patients. Carfilzomib–lenalidomide–dexamethasone plus autologous stem cell transplantation treatment improved the adverse effect of Gain1q but not Amp1q. Transcriptomic data showed that additional 1q copies were associated with deregulation in apoptosis signaling, p38 MAPK signaling, and Myc-related genes.
Subject terms: Risk factors, Myeloma, Myeloma
Introduction
Multiple myeloma (MM) is a common hematological neoplasia, and in the last 2 decades, a meaningful survival improvement has been reached thanks to the introduction of novel drugs, with nowadays more than 60% of transplant-eligible patients alive 8 years from initial diagnosis [1]. Nevertheless, despite this improvement, there is still a subset of patients with a reduced benefit from new treatment approaches and a dismal outcome. These patients with “high risk” disease represent a very heterogeneous group, including patients with high tumor burden, genomic and cytogenetic alterations, gene expression profiles, or presence of circulating tumor cells and extramedullary disease [2].
The genomic and cytogenetic events that occur in the genesis, evolution, and progression of MM have been thoroughly investigated [3, 4]. Recurrent chromosomal abnormalities that can be detected by interphase fluorescent in situ hybridization (iFISH) allowed a classification of MM patients based on cytogenetic events, with del(17p), t(4;14) and t(14;16) classically considered as high-risk by IMWG [5].
Indeed, additional copies of chromosome 1q are one of the most frequent chromosomal abnormalities, reported in precursor stages, found in 30–40% of newly diagnosed (ND) MM patients [6] and 50–80% of relapsed-refractory patients [7]. The clonal size [8] of 1q copies seems to correlate with the prognosis. Moreover, the prognostic value of additional copies of 1q21 may depend on the presence of other primary genetic events [9]. Several reports, including clinical trials and real-world data, correlate the presence of 1q abnormalities with adverse outcomes [10–16]. Based on these data, 1q alterations have been incorporated in recent staging systems for NDMM [17–19]. As an example, 1q21+ has recently been included in the R2-ISS [18], and other analyses investigated the prognostic impact of Amp1q (at least four copies of 1q) vs. Gain1q (three copies of 1q) [15, 19–21].
The possible role of specific novel treatment approaches in reducing the adverse outcomes related to high-risk features is currently under evaluation. Response rates are similar in patients with or without 1q alterations, but progression-free survival (PFS) and overall survival (OS) are inferior in patients with 1q abnormalities vs not if patients are treated with autologous stem cell transplantation (ASCT) [15], immunomodulatory drugs (IMID) or bortezomib [14, 21].
Few data are available yet on the impact of Carfilzomib in transplant-eligible NDMM patients with 1q abnormalities.
Hyperexpression of genes located in the 1q region has also been associated with adverse prognosis [22]. The adverse outcome related to 1q copy number and 1q clonal size suggests a “dose effect”, where several genes located on chromosome 1q could be involved (e.g. CKS1B, PSMD4, ADAR1, MCL-1); nevertheless, it is not clear if 1q alteration could mirror genetic instability rather than be the real driver of poor prognosis [23]. The identification of candidate key genes and pathways that are behind 1q abnormalities could potentially pave the way for new therapeutic targets.
The primary aim of our analysis is to evaluate the impact of Amp1q vs Gain1q on outcomes in patients enrolled in the randomized FORTE clinical trial (NCT02203643) in order to describe their impact in a carfilzomib-based therapy setting on the background of the established role of Gain/Amp1q as a broad prognostic factor. Our secondary aim is to understand the transcriptomic changes in Amp1q, Gain1q, and Normal 1q that could elucidate the biological drivers of the adverse outcome related to Amp1q/Gain1q.
Methods
Study design and participants
ND transplant-eligible MM patients aged 65 years or younger were enrolled between February 23, 2015 and April 5, 2017. Patients were randomized (R1) into three induction/intensification/consolidation arms: the KCd-ASCT arm included four 28-day induction cycles with KCd, melphalan 200 mg/sqm followed by ASCT (MEL200-ASCT), and four KCd consolidation cycles. the KRd12 arm consisted of twelve 28-day cycles with KRd without upfront MEL200-ASCT; Patients in the KRd-ASCT arm received four 28-day induction cycles with KRd, MEL200-ASCT, and four KRd consolidation cycles.
After consolidation, a second randomization (R2) to two maintenance regimens was performed in eligible patients. Patients in the KR maintenance arm received carfilzomib for up to 2 years and lenalidomide until progression, while patients in the R maintenance arm received lenalidomide alone until progression. Full details on the study treatment were previously described [24].
The FORTE trial was approved by the ethics or Institutional Review Boards at each of the participating centers. All patients gave written informed consent before entering the study, which was performed in accordance with the Declaration of Helsinki and the Good Clinical Practice guidelines.
Cytogenetic risk evaluation
Cytogenetic risk was assessed by a centralized laboratory receiving bone marrow aspirate samples collected during the screening phase. Chromosome abnormalities (CA) were assessed by FISH analysis performed on CD138+ purified bone marrow plasma cells (BMPCs). BMPCs were enriched using anti-CD138-coated magnetic microbeads and an AutoMACS Pro separator (Miltenyi Biotech) following the manufacturer’s instructions [25]. The FISH analysis included probes to detect conventionally defined high-risk disease [t(4;14) and/or t(14;16) and/or del(17p)] [26] and Gain/Amp1q as well (LPH039 probe, Cytocell™, Oxford Gene Technology, Oxford, England).
Two hundred BMPC nuclei from each sample were counted and scored. The cut-off value for CA positivity was 15% for translocations and 10% for copy number alterations. Cut-offs were defined according to the mean ± 3 standard deviations of abnormal signals detected in the bone marrow plasma cells of healthy donors [25, 27].
Patients were defined as Gain(1q) positive if ≥10% of nuclei with ≥3 copies of 1q were detected and the definition of Amp(1q) was not met.
Patients were defined as Amp(1q) positive if ≥20% of nuclei with ≥4 copies of 1q21 were detected. The 20% cut-off for Amp(1q) positivity was recommended by FISH guidelines [27] and consistent with the one applied by other groups [14, 28]. The reliability of the cut-off was further verified using an Epanechnikov kernel-smoothed estimated hazard rate to study the risk of progression or death according to the % of nuclei with ≥4 copies of 1q (Fig. S1).
MRD evaluation
A centralized evaluation of minimal residual disease (MRD) by multiparameter flow cytometry (MFC; sensitivity threshold of 10-5) [24] was performed before maintenance and thereafter every 6 months during maintenance in all patients achieving at least a very good partial response (VGPR). Data were analyzed on intention-to-treat (ITT), with patients with a positive MRD test result or who had not undergone MRD testing (either because they missed evaluation, or they achieved < VGPR) considered MRD positive. The calculation of 1-year sustained MRD negativity is described in the supplementary appendix.
RNA sequencing data generation and analysis
Data from patients enrolled in the prospective observational Multiple Myeloma Research Foundation (MMRF) CoMMpass study (NCT01454297) were used as a test set, and data from additionally sequenced patients enrolled in the FORTE trial were used as a validation set.
The Interim Analysis (IA)14 release of CoMMpass was used for the analyses, 1q copy number abnormalities (CNAs) in the CoMMpass patients were defined using molecular data with a technique (SeqFISH) that was previously cross-validated against conventional FISH [29]. RNAseq data in the CoMMpass study were generated as previously described [30].
Data deposition, RNAseq data analysis, and RNAseq data generation in the additionally sequenced patients enrolled in the FORTE trial are described in the Supplementary appendix.
Statistical analysis
The efficacy analysis included patients in the ITT population who met the biomarker criteria for risk assessment (1q probe analyzed by FISH and number of 1q copies available), whereas patients with incomplete data about 1q copy number were excluded from this analysis. Patients with evaluable 1q copy numbers were stratified into three groups according to the presence of Amp(1q), Gain(1q), or no 1q abnormalities (Normal 1q) as defined above. The aim of this analysis was to compare the outcome of patients with Amp(1q), Gain(1q), and Normal 1q.
This analysis was a post-hoc analysis that was not prespecified in the protocol.
PFS was calculated from the date of randomization (R1 or R2, according to the endpoint) to the date of first observation of progressive disease (PD) or death from any cause. Patients who did not progress or were lost to follow-up, or who withdrew from the trial were censored at the time of the last disease assessment.
OS was calculated from the date of first randomization to the date of death from any cause.
Time-to-event data were analyzed using the Kaplan–Meier method. The Cox proportional hazards model was used to estimate the HR values and the 95% confidence intervals (CIs). Cox models were adjusted for Revised-International Staging System (R-ISS) stage (R-ISS III vs. II vs. I vs. not available), age (≥60 vs. <60 years), and randomization arm (KRd-ASCT vs. KRd12 vs. KCd-ASCT). The significance level was set at P < 0·05.
The statistical analyses were performed using R (v4.0·2). The data cut-off point was January 7, 2021.
Role of the funding source
The UNITO-MM-01/FORTE trial was sponsored by the Università degli Studi di Torino (Turin, Italy), Department of Molecular Biotechnology and Health Sciences. Amgen and Celgene/Bristol Myers Squibb provided an unrestricted grant to conduct the trial but had no role in study design, data collection, data analysis, data interpretation, writing of the report, or the decision to submit the manuscript for publication. International Myeloma Society (IMS) supported the research analyses presented in this paper with an award conferred to the principal investigator of the trial.
Results
Study population
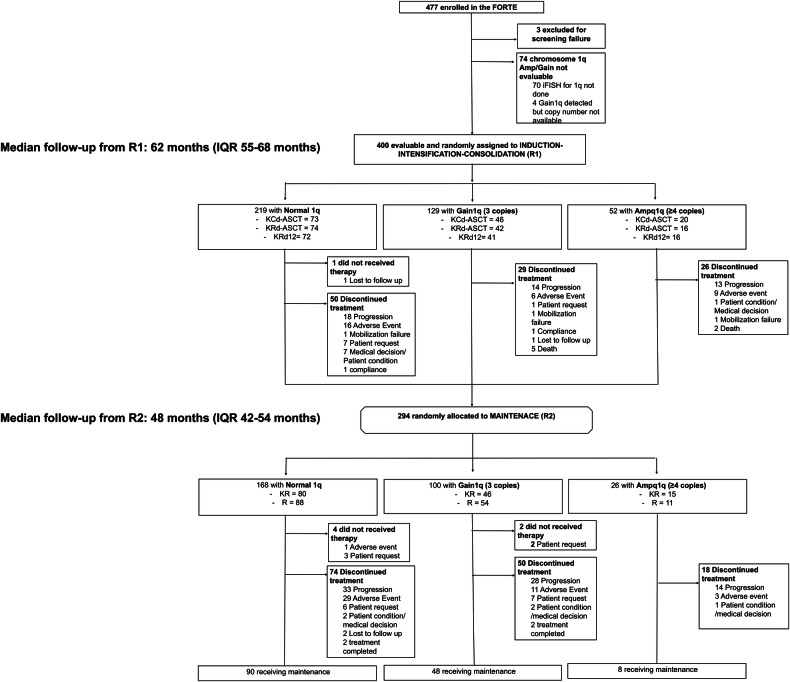
A total of 477 patients were enrolled in the FORTE trial and 474 were randomized into one of the three induction–intensification–consolidation arms (R1). In 74 patients, the evaluation of 1q abnormalities was missing, or 1q copy number was not available (Fig. 1). Of 400 patients evaluable for 1q abnormalities (ITT 1q population), 52 (13%) had Amp1q, 129 (32%) Gain1q and 219 (55%) had Normal 1q.
Fig. 1.
Consort diagram of patients enrolled in the FORTE trial.
Regarding group disposition, 26 patients did not meet the definition of Amp1q but had a borderline number of nuclei with ≥4 copies of 1q (10–19% of total cells). All these 26 patients were not classified as Amp1q, but they were classified as Gain1q because they had a mean number of nuclei with three copies of 59% (range 33–78%) and a mean number of nuclei with ≥3 copies (3 copies + ≥4 copies) of 73% (range 48–91%).
Median follow-up from the first randomization was 62 months [Interquartile range (IQR) 55–68 months].
Demographic features and treatment were well balanced between patients with Amp1q, Gain1q, and Normal1q. High-risk disease features (ISS stage II–III, high-risk cytogenetics, and high LDH) were enriched in the group of patients with 1q abnormalities compared to Normal 1q with no statistically significant differences between patients with Amp1q vs. Gain1q, except for an increased proportion of patients with high LDH (p = 0.0024) in Amp1q group (Table 1). Of note, patients with hyperdiploid status were less represented in Amp1q cases (23% vs. 36% vs. 45% in Amp1q vs. Gain1q vs. Normal1q cases, p = 0.016).
Table 1.
Patient characteristics of patients randomized for induction therapy (R1) in the FORTE trial according to 1q21 status.
| FORTE population | 1q Amp/gain not evaluablea | Amp1q | Gain1q | Normal 1q | p-valueb | ||
|---|---|---|---|---|---|---|---|
| N = 474 | N = 74 | N = 52 | N = 129 | N = 219 | |||
| Age | Median (IQR) | 57- (51–62) | 55- (50–61) | 59 – (53–62) | 58 – (53–63) | 57- (51–62) | 0.6693 |
| ISS | I | 240 (51) | 50 (68) | 15 (29) | 61 (47) | 114 (52) | 0.0764 |
| II | 152 (32) | 15 (20) | 23 (44) | 41 (33) | 73 (33) | ||
| III | 82 (17) | 9 (12) | 14 (27) | 27 (15) | 32 (15) | ||
| Cytogenetic abnormalities | Standard risk | 275 (67) | 5 (56) | 25 (48) | 74 (57) | 171 (78) | 0.3223 |
| High riskc | 133 (33) | 4 (44) | 27 (52) | 55 (43) | 47 (22) | ||
| Missing | 66 | 65 | 0 | 0 | 1 | ||
| del(17p) | 58 (15) | 0 | 11 (21) | 15 (12) | 32 (15) | 0.1064 | |
| t(4;14) | 65 (16) | 0 | 17 (33) | 31 (24) | 17 (8) | 0.2656 | |
| t(14;16) | 21 (5) | 2 (3) | 7 (13) | 11 (9) | 2 (1) | 0.4097 | |
| LDH | Normal | 397 (87) | 68 (93) | 31 (65) | 108 (86) | 190 (90) | 0.0024 |
| High | 61 (13) | 5 (7) | 17 (35) | 17 (14) | 22 (10) | ||
| Missing | 16 | 1 | 4 | 4 | 7 | ||
| Induction Therapy (R1) | KCd-ASCT | 159 (34) | 20 (27) | 20 (38) | 46 (36) | 73 (33) | 0.9360 |
| KRd-ASCT | 157 (33) | 28 (38) | 16 (31) | 41 (32) | 72 (33) | ||
| KRd-12 | 158 (33) | 26 (35) | 16 (31) | 42 (33) | 74 (34) |
Percentage may not total 100 because of rounding. % calculated on the available patients.
IQR interquartile range, ISS International Staging System, LDH lactate dehydrogenase; FISH fluorescence in situ hybridization, K carfilzomib, R lenalidomide, d dexamethasone, KCd-ASCT: KCd induction-ASCT-KCd consolidation, KRd-ASCT KRd induction-ASCT-KRd consolidation, KRd12 12 cycles of KRd.
a70/474: 1q CNA not done, 4/474: Gain1q detected but copy number not available.
bAmp1q vs. Gain1q comparison.
cHigh risk-cytogenetic abnormalities: defined as del(17p) and/or t(4;14) and/or t(14;16).
ITT analysis
In the ITT analysis, rate of premaintenance ≥ VGPR was 77% (95% CI 63–87%) in Amp1q, 84% (95% CI 76–90%) in Gain1q and 84% (95% CI 78–88%) in Normal 1q (p = 0.49); premaintenance ≥ CR was achieved in 46% (95% CI 32–61%), 47% (95% CI 38–55%) and 52% (95% CI 45–58%) of patients, respectively (p = 0.59). Similarly, the rate of premaintenance 10−5 MRD negativity MFC was comparable in Amp1q [44% (95% CI 30–59%)], Gain1q [55% (95% CI 46–64%)], and Normal 1q [55% (95% CI 48–62%)] (p = 0.35). There was a trend towards a lower 1-year sustained MRD negativity rate in Amp1q [19% (95% CI 10–33%)] vs Gain1q [32% (95% CI 24–41%)] and Normal 1q [37% (95% CI 31–44%)] (p = 0.05) (Table S1).
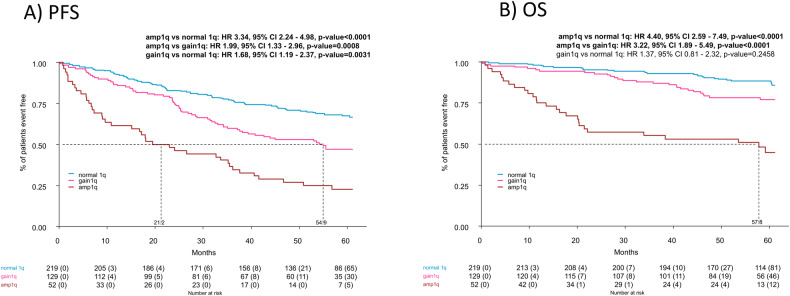
PFS from R1 (Fig. 2A) was significantly inferior in the presence of Amp1q vs. Normal1q (HR 3.34, 95% CI 2.24–4.98, p < 0.0001) and Amp1q vs. Gain1q (HR 1.99, 95% CI 1.33–2.96, p = 0.0008). Patients with Gain1q had also a significantly shorter PFS compared with Normal 1q (HR 1.68, 95% CI 1.19–2.37, p = 0.0031). 4-year PFS rate was 27% (95% CI 17–42%)in Amp1q, 53% (95% CI 45–63%) in Gain1q and 71% (95% CI 65–77%) in Normal 1q.
Fig. 2. Outcomes according to 1q copies.
Progression-free survival (A) and overall-survival (B) from first randomization (R1) according to 1q subgroups. HR values and the 95% CIs were estimated with a Cox proportional hazards model adjusted for the R-ISS stage (R-ISS III vs. II vs. I vs. not available), age (≥60 vs. <60 years), and randomization arm (KRd-ASCT vs. KRd12 vs. KCd-ASCT).
We ran an additional analysis using a more conservative cut-off to define Gain1q (20% instead of 10%). Only 19 patients were reclassified from Gain1q positive to Normal 1q, and the results were comparable to what was observed in the main analysis using a 10% cut-off (Supplementary appendix).
The presence of Amp1q was also associated with significantly shorter OS compared to Normal 1q (HR 4.40, 95% CI 2.59–7.49, p < 0.0001) and to Gain1q (HR 3.22, 95% CI 1.89–5.49, p < 0.0001 (Fig. 2B), with no significant differences between Gain1q and Normal1q groups. 4-year OS rate was 53% (95% CI 41–69%) Amp1q, 78% (95% CI 71–86%) Gain1q and 91% (95% CI 87–95%) in Normal1q.
294 patients of the ITT 1q population were randomized to maintenance (R2): 26/52 (50%) with Amp1q, 100/129 (78%) with Gain1q and 168/219 (77%) with Normal 1q. Reasons for discontinuation before R2 are listed in Fig. 1; there was a higher rate of discontinuation due to PD in the Amp1q group (13/52, 25%) compared to Gain1q (14/129, 11%) and Normal 1q (18/219, 8%). Patients’ characteristics of ITT 1q patients randomized to maintenance are summarized in the Supplementary Appendix (Table S2).
The PFS from second randomization (R2) was significantly inferior in patients with Amp1q vs. Normal1q (HR 3.05, 95% CI 1.69–5.51, p = 0.0002) and Gain1q vs. Normal 1q (HR 1.81, 95% CI 1.14–2.86, p = 0.0116) while only a trend was found in Amp1q vs. Gain1q (HR 1.69, 95% CI 0.93–3.06, p = 0.0858) at the current follow-up (Fig. S2). 3-year PFS rate was 46% (95% CI 30–70%) vs. 65% (95% CI 56–75%) vs. 80% (95% CI 74–86%) in Amp1q vs. Gain1q vs. Normal 1q groups.
Subgroup analyses
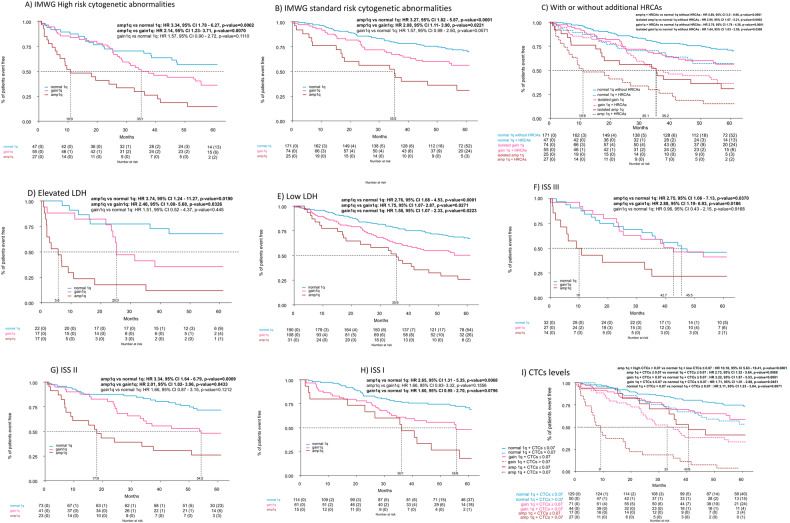
Subgroup analyses for PFS confirmed the negative prognostic impact of Amp1q vs. Gain1q and Normal q1 in all subgroups of patients (Fig. S3). Of interest, the co-occurrence of Amp1q with other baseline risk features (Fig. 3) identifies a group of patients with an extremely poor outcome [median PFS of high-risk cytogenetics + Amp1q 10.9 months (95% CI 7.3–36.1); High LDH + Amp1q 5.6 months (95% CI 1.9–17.9); ISS3 + Amp1q 10 months (95% CI 5.8–NR)]. Interestingly, Amp1q without additional high-risk chromosomal abnormalities (HRCA) had a similar outcome compared with Gain1q with additional HRCA [Fig. 3C, median PFS 35.2 months (95% CI 17.1–NR) vs. 35.1 months (95% CI 28.4–NR); HR 1.30, 95% CI 0.54–3.14, p = 0.56]. However, the worst outcome was detected in Amp1q patients with additional HRCA, suggesting that there is an additive effect of additional HRCA even in the context of very high-risk abnormalities like Amp1q.
Fig. 3. Progression-free survival in subgroups of interest.
Progression-free survival from first randomization (R1) according to 1q alterations and concomitant cytogenetic risk (A–C), LDH levels (D and E), ISS (F–H), and circulating tumor cells (CTCs) levels (I).
We further analyzed the relation of Gain1q and Amp1q with a novel prognostic marker described in the FORTE cohort, namely the levels of baseline circulating tumor cells (CTCs) in the peripheral blood with a cutoff of 0.07% by multiparametric flow cytometry [31]. As with other baseline risk features, the coexistence of Amp1q and high CTCs led to the worst PFS [median PFS 9 months (95% CI 6.3–18.1), Fig. 3I].
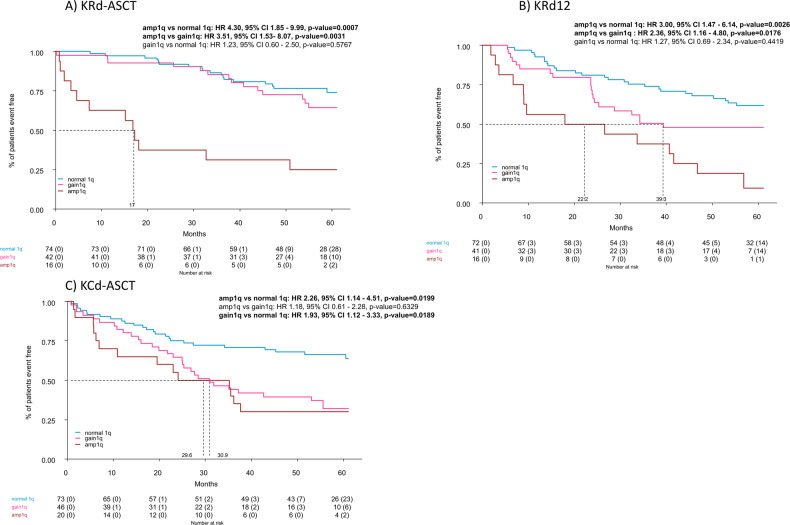
Regarding treatment, the negative impact of Amp1q was evident in all treatment arms (Figs. 4 and S3); for what concerns Gain1q, patients treated with KRd-ASCT arm showed a superimposable PFS compared to Normal1q [4 year PFS 72% (95% CI 60–88%) for Gain1q vs. 77% (95% CI 67–87%) for Normal1q; HR 1.23, 95% CI 0.60–2.50, p = 0.5777]. PFS of Gain1q vs Normal 1q was still inferior in patients treated with KCd-ASCT [4-y PFS: 39% (95% CI 27–57%) vs. 68% (95% CI 58–80%), HR 1.93, 95% CI 1.12–3.33, p = 0.0189], while in patients treated with KRd12 a non-significant trend was noted [4-y PFS: 48% (95% CI 34–67%) vs. 68% (95% CI 58–80%), HR 1.27, 95% CI 0.69–2.34, p = 0.4419].
Fig. 4. Progression-free survival from first randomization (R1) according to treatment received and 1q alterations.
A KRd-ASCT, B KRd12, and C KCd-ASCT.
Of note, the achievement of premaintenance MRD negativity by MFC 10−5 improved the outcome of all patients’ groups (Fig. S4B); nevertheless, Ampl1q still retained its prognostic significance (Fig. S4B). In patients achieving 1-year sustained MRD negativity, no significant PFS differences can be found according to 1q subgroups at current follow-up (Fig. S4C). On the other hand, Amp1q patients not achieving MRD negativity have a median PFS of only 7.3 months (95% CI 5.8–17.1), representing a very high-risk population (Fig. S4A).
RNAseq of malignant plasma cells according to 1q copies
In order to compare groups with a similar sample size, in the ANOVA-like analysis of RNAseq data according to 1q copies [32], we randomly selected a population of patients with Normal 1q and Gain1q to match the size of the Amp1q group (analyzed patients are listed in Table S3).
Patients with a concomitant t(4;14) translocation were excluded from the analysis because the high expression of FGFR3 in these cases interacted with the clustering of 1q-defined subgroups. Indeed, in our case series, this effect was pronounced in Amp1q cases, where patients with the cooccurrence of t(4;14) clustered together (Fig. S5A).
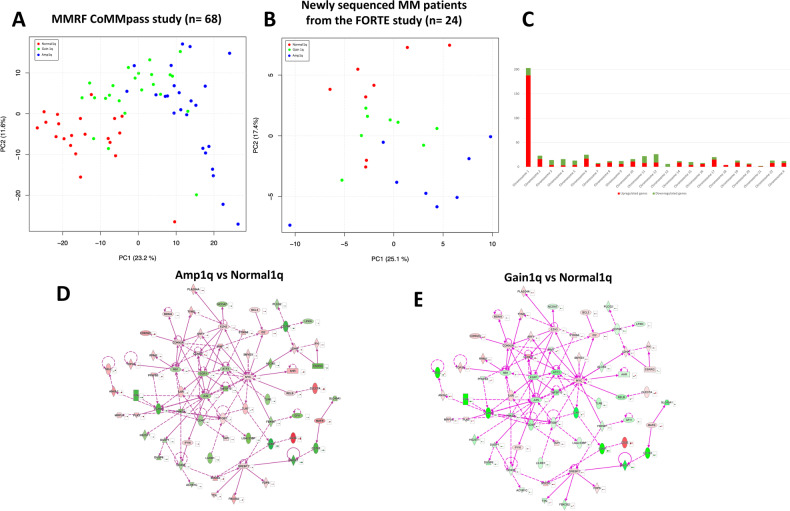
An ANOVA-like analysis of RNAseq data of 68 patients enrolled in the CoMMpass study (Table S3) showed 498 differentially expressed genes that were able to separate 1q-defined groups in a Principal Component Analysis (Fig. 5A). Clustering according to 1q groups is shown in Fig. S5B. The list of differentially expressed genes in Amp1q, Gain1q, and Normal1q is provided in Table S4. The identified genes were able to separate 1q-defined groups also in an independent population of 24 patients enrolled in the FORTE trial that were not included in the CoMMpass study (Fig. 5B).
Fig. 5. Transcriptomic deregulation according to 1q copies.
Principal component analysis using RNAseq data was able to separate NDMM patients according to 1q copy numbers in the MMRF CoMMpass study (A). The separation was maintained when applied to a validation cohort of newly sequenced NDMM patients from the FORTE trial (B). Number of differentially expressed genes divided according to chromosome number are presented in Panel C. Ingenuity Pathway connectivity analysis of differentially expressed genes in Amp1q vs. Normal1q (D) and Gain1q vs. Normal1q (E) shows a deregulated gene network centered on Myc. Green color indicates down-regulation and red color up-regulation.
Of note, many (203/498) but not all differentially expressed genes (DEGs) were located in chromosome 1 (Fig. 5C). Moreover, 188 of the 203 DEGs in chromosome 1 were upregulated consistently with the additional 1q copies.
An ingenuity pathways analysis showed that apoptosis signaling (z-score 2.646) and p38 MAPK signaling pathways (z-score 2.646) were the most deregulated pathways in Amp1q patients (Table S5).
Ingenuity pathway connectivity analysis of differentially expressed genes in Amp1q vs. Normal1q (Fig. 5D) and Gain1q vs. Normal1q (Fig. 5E) showed a deregulated gene network that was centered on the Myc gene.
Notably, genes that were deregulated in Gain1q cases, have usually a more pronounced deregulation in Amp1q cases (e.g. EGR1, Fig. S5C). However, some genes (e.g. AHR and RELB, Fig. S5C) showed a change in expression direction in Gain1q vs. Amp1q cases.
Of note, CD55, a gene located in chromosome 1q, which is known to promote resistance to complement-dependent cytotoxicity induced by anti-CD38 monoclonal antibodies (e.g. daratumumab) [33], was upregulated in Gain and especially Amp1q cases. (Fig. S5C).
Moreover, the GPRC5D gene, a relevant immunotherapeutic target in MM [34], despite being located outside of chromosome 1q, was upregulated in Gain and especially Amp1q patients in our analysis (Fig. S5C).
Discussion
We demonstrated that the number of 1q copies detected by FISH is an independent prognostic marker in a population of transplant-eligible NDMM patients treated with carfilzomib-based combinations.
Amp1q (≥4 copies of 1q) was associated with a very poor PFS and OS compared with Normal but also with Gain1q, while patients with Gain1q (3 copies of 1q), showed an intermediate outcome.
The different prognostic impact of Gain1q and Amp1q is in line with some studies [9, 14, 16], while other reports showed similar OS [15, 21, 35].
Technical variability in calling 1q status, differences in follow-up time, differences in treatment strategies, and a different distribution of concomitant high-risk features may account for some of these discrepancies.
No consensus exists on the optimal cutoffs to define Gain1q vs. Amp1q. Here we used a 20% cut-off of nuclei with ≥4 copies of 1q to define Amp1q positivity, and an Epanechnikov kernel-smoothed estimated hazard rate showed that the risk of progression is higher if the clonal size of cells with ≥4 copies of 1q is higher.
While using a more stringent cut-off may identify a smaller subset of patients with a higher risk of progression, a recent report showed that even the presence of small 1q positive subclones identified through single-cell technology, may affect prognosis [36]. Indeed, the selection of 1q subclones emerging at relapse may show a similar negative impact on PFS and OS compared to patients who have the alteration from diagnosis [36].
Defining the prognostic impact of Amp 1q vs. Gain1q is meaningful if we can also identify subgroups of patients who can benefit from different treatment approaches. In a retrospective analysis of patients treated with bortezomib–lenalidomide–dexamethasone (VRd), Amp1q predicted a poor PFS regardless of other cytogenetic abnormalities, while patients with Gain1q had a dismal outcome only when Gain1q was associated with t(4;14), or t(14;16) or del (17p) [14].
In the E1A11 trial, standard risk NDMM patients not intended for upfront transplant were randomized to VRd vs. KRd induction followed by indefinite or 2-year R maintenance. Since the enrollment of t(4;14), del(1p), and Gain/Amp1q patients were allowed, a post-hoc analysis focusing on chromosome 1 alterations was performed [37]. Gain1q and Amp1q patients did show poor PFS with either VRd or KRd [37]. However, looking at OS, KRd seemed to overcome the negative impact of Gain1q, but not Amp1q.
In our study, both Gain1q and especially Amp1q showed a lower PFS compared to patients with Normal 1q, independently from the presence of other high-risk features. Nevertheless, treatment with KRd induction-ASCT-KRd consolidation seemed to overcome the negative prognostic value of Gain1q. Amp1q remained strongly associated with poor prognostic in all treatment arms, and many of these patients did not reach the maintenance phase due to early progression. Moreover, when Amp1q was associated with other high-risk features (e.g. other high-risk cytogenetic abnormalities, high LDH, ISS 3, high CTCs) or when patients did not reach bone marrow MRD negativity, PFS was <12 months, clearly identifying a patient population with a very high unmet medical need. Indeed, despite isolated Amp1q already predicted an adverse outcome, there is a clear additive effect of additional poor prognostic factors even in Amp1q-positive patients.
Achieving and sustaining over time MRD negativity was the only factor ameliorating the prognosis of Amp1q patients, indicating the need to explore novel treatment options to pursue sustained MRD in these patients, such as early intensification of treatment in patients harboring Amp1q not reaching an MRD negativity or not sustaining it over time.
We analyzed RNAseq to detect DEGs associated with Gain1q and Amp1q that can shed light on druggable targets/pathways to be further tested in this high-risk patient population.
The concept that copy number alterations can cause a functional impact on the transcriptome has been shown before [38]. In that study, 1q alterations were associated with the greatest impact on gene expression, deregulating pathways related to cell cycle, proliferation, and expression of immunotherapy targets [38].
In our study, we found that deregulation in apoptosis signaling and p38 MAPK signaling pathways and a gene network centered on Myc may contribute to the high-risk behavior associated with additional 1q copies.
Moreover, we found in Amp1q cases, an upregulation of CD55 that may reduce the efficacy of anti-CD38 monoclonal antibodies relying on complement-dependent cytotoxicity as the main mechanism of action. An upregulation of GPRC5D can be found in Amp1q cases as well in our cohort and bispecific antibodies [39]/CAR T cells [40] targeting GPRC5D are under evaluation in clinical trials.
Subgroup analyses on Amp1q patients treated with these agents are warranted to understand if the transcriptomic results translate into a different clinical benefit with these therapies.
The exclusion from the transcriptomic analysis of patients with concomitant t(4;14) may have reduced the risk of cluster bias. However, though we have not observed a clear effect of other concomitant alterations, the impact of the coexistence of other alterations cannot be completely ruled out. Indeed, the main limitation of our analysis is that we used bulk RNAseq, while single-cell RNAseq may identify more precisely the transcription alterations in 1q positive cells. In a single-cell RNA seq study addressing +1q cells, an upregulation of mitochondrial oxidative phosphorylation causing increased reactive oxygen species and reduced energy stress, compared with cells without +1q, was found [41]. Using single-cell RNAseq, the MYC gene expression signature is enriched amongst +1q samples, with MYC cooperating with MCL1 to enrich the MYC gene expression signature in +1q samples. This is indeed consistent with our analysis using bulk RNAseq data.
In conclusion, in our cohort, Amp1q identifies patients with very high-risk MM, while Gain1q patients are at intermediate risk of progression and/or death and may benefit from the KRd-ASCT-KRd approach. Additional copies of 1q induced relevant transcriptomic changes in MM cells, and it may be worth exploring the use of specific agents in this patient subset.
Supplementary information
Acknowledgements
The authors wish to thank all the study participants, caregivers, physicians, nurses, and data managers of the participating centers. The UNITO-MM-01/FORTE trial was sponsored by the Università degli Studi di Torino (Italy), Department of Molecular Biotechnology and Health Sciences. Amgen and Celgene/Bristol Myers Squibb provided an unrestricted grant to conduct the trial but had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or publication of this contribution. The authors were not precluded from accessing data in the study, and they accept responsibility for submitting this manuscript for publication. International Myeloma Society (IMS) supported the research analyses presented in this paper with an award conferred to the principal investigator of the trial (FG).
Author contributions
All authors contributed to the acquisition, analysis, or interpretation of data for this work. All authors critically reviewed the work for important intellectual content, approved the final version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors had access to and verified the underlying data. MD, LB, MR, BB, MB, and FG contributed to the conceptualization, formal analysis, methodology, and writing of the original draft. MD and LB contributed to the visualization. MB and FG contributed to supervision. All authors contributed to data curation, investigation, resources, validation, and writing of the manuscript in terms of review and editing. All authors had access to all the data reported in the study and had final responsibility for the decision to submit this manuscript for publication.
Data availability
After the publication of this article, data collected for this analysis and related documents will be made available to others upon reasonably justified request, which needs to be written and addressed to the attention of the corresponding author FG at the following e-mail address: francesca.gay@unito.it. The sponsor of the UNITO-MM-01/FORTE trial, the University of Torino (Italy), via the corresponding author FG, is responsible for evaluating and eventually accepting or refusing every request to disclose data and their related documents, in compliance with the ethical approval conditions, in compliance with applicable laws and regulations, and in conformance with the agreements in place with the involved subjects, the participating institutions, and all the other parties directly or indirectly involved in the participation, conduct, development, management and evaluation of this analysis.
Competing interests
MD has received honoraria for lectures for GlaxoSmithKline, Sanofi, and Janssen; has served on the advisory boards for GlaxoSmithKline, Sanofi, and Bristol Myers Squibb. AB has served on the advisory boards for Amgen, GlaxoSmithKline, Janssen, Pfizer. PG has served on the advisory boards for Takeda and Incyte. EZ recieved honoraria from Janssen, Bristol-Myers Squibb, Amgen, Takeda. BB has received honoraria from Amgen, Janssen, Novartis, BeiGene, Bristol Myers Squibb, GlaxoSmithKline, Jazz pharmaceuticals, Astrazeneca and Incyte; has served on the advisory boards for Amgen and Jazz pharmaceuticals. MB has received honoraria from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol Myers Squibb, and AbbVie; has served on the advisory boards for Janssen and GlaxoSmithKline; has received research funding from Sanofi, Celgene, Amgen, Janssen, Novartis, Bristol Myers Squibb, and Mundipharma. PM has received honoraria from Celgene, Janssen, Takeda, Bristol Myers Squibb, Amgen, Novartis, Gilead, Jazz, Sanofi, AbbVie, and GlaxoSmithKline; has served on the advisory boards for Celgene, Janssen, Takeda, Bristol Myers Squibb, Amgen, Jazz, Sanofi, AbbVie, and GlaxoSmithKline. FG has received honoraria from Amgen, Celgene, Janssen, Takeda, Bristol Myers Squibb, AbbVie, and GlaxoSmithKline; has served on the advisory boards for Amgen, Celgene, Janssen, Takeda, Bristol Myers Squibb, AbbVie, GlaxoSmithKline, Roche, Adaptive Biotechnologies, Oncopeptides, bluebird bio, and Pfizer. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01075-x.
References
- 1.Perrot A, Lauwers-Cances V, Cazaubiel T, Facon T, Caillot D, Clement-Filliatre L, et al. Early versus late autologous stem cell transplant in newly diagnosed multiple myeloma: long-term follow-up analysis of the IFM 2009 trial. Blood. 2020;136:39. doi: 10.1182/blood-2020-134538. [DOI] [Google Scholar]
- 2.Caro J, Al Hadidi S, Usmani SZ, Yee AJ, Raje N, Davies FE. How to treat high-risk myeloma at diagnosis and relapse. In: American Society of Clinical Oncology, editor. Educational book. Alexandria (USA); vol. 2021. [DOI] [PubMed]
- 3.Kumar SK, Rajkumar SV. The multiple myelomas—current concepts in cytogenetic classification and therapy. Nat Rev Clin Oncol. 2018;15:409–21. doi: 10.1038/s41571-018-0018-y. [DOI] [PubMed] [Google Scholar]
- 4.Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14:100–13. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J Clin Oncol. 2015;33:2863–9. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonseca R, Van Wier SA, Chng WJ, Ketterling R, Lacy MQ, Dispenzieri A, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–40. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 7.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: Incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–32. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An G, Li Z, Tai YT, Acharya C, Li Q, Qin X, et al. The impact of clone size on the prognostic value of chromosome aberrations by fluorescence in situ hybridization in multiple myeloma. Clin Cancer Res. 2015;21:2148–56. doi: 10.1158/1078-0432.CCR-14-2576. [DOI] [PubMed] [Google Scholar]
- 9.Locher M, Steurer M, Jukic E, Keller MA, Fresser F, Ruepp C, et al. The prognostic value of additional copies of 1q21 in multiple myeloma depends on the primary genetic event. Am J Hematol. 2020;95:1562–71. doi: 10.1002/ajh.25994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avet-Loiseau H, Attal M, Campion L, Caillot D, Hulin C, Marit G, et al. Long-term analysis of the ifm 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J Clin Oncol. 2012;30:1949–52. doi: 10.1200/JCO.2011.36.5726. [DOI] [PubMed] [Google Scholar]
- 11.Nemec P, Zemanova Z, Greslikova H, Michalova K, Filkova H, Tajtlova J, et al. Gain of 1q21 is an unfavorable genetic prognostic factor for multiple myeloma patients treated with high-dose chemotherapy. Biol Blood Marrow Transplant. 2010;16:548–54. doi: 10.1016/j.bbmt.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Shah GL, Landau H, Londono D, Devlin SM, Kosuri S, Lesokhin AM, et al. Gain of chromosome 1q portends worse prognosis in multiple myeloma despite novel agent-based induction regimens and autologous transplantation. Leuk Lymphoma. 2017;58:1823–31. doi: 10.1080/10428194.2016.1260126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varma A, Sui D, Milton DR, Tang G, Saini N, Hasan O, et al. Outcome of multiple myeloma with chromosome 1q gain and 1p deletion after autologous hematopoietic stem cell transplantation: propensity score matched analysis. Biol Blood Marrow Transplant. 2020;26:665–71. doi: 10.1016/j.bbmt.2019.12.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt TM, Barwick BG, Joseph N, Heffner LT, Hofmeister CC, Bernal L, et al. Gain of Chromosome 1q is associated with early progression in multiple myeloma patients treated with lenalidomide, bortezomib, and dexamethasone. Blood Cancer J. 2019;9:94. doi: 10.1038/s41408-019-0254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdallah N, Greipp P, Kapoor P, Gertz MA, Dispenzieri A, Baughn LB, et al. Clinical characteristics and treatment outcomes of newly diagnosed multiple myeloma with chromosome 1q abnormalities. Blood Adv. 2020;4:3509–19. doi: 10.1182/bloodadvances.2020002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neben K, Lokhorst HM, Jauch A, Bertsch U, Hielscher T, van der Holt B, et al. Administration of bortezomib before and after autologous stem cell transplantation improves outcome in multiple myeloma patients with deletion 17p. Blood. 2012;119:940–8. doi: 10.1182/blood-2011-09-379164. [DOI] [PubMed] [Google Scholar]
- 17.Abdallah NH, Binder M, Rajkumar SV, Greipp PT, Kapoor P, Dispenzieri A, et al. A simple additive staging system for newly diagnosed multiple myeloma. Blood Cancer J. 2022;12:1–9. doi: 10.1038/s41408-022-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Agostino M, Cairns DA, Lahuerta JJ, Wester R, Bertsch U, Waage A, et al. Second Revision of the International Staging System (R2-ISS) for overall survival in multiple myeloma: a European Myeloma Network (EMN) report within the HARMONY Project. J Clin Oncol. 2022;40:3406–18. doi: 10.1200/JCO.21.02614. [DOI] [PubMed] [Google Scholar]
- 19.Perrot A, Lauwers-Cances V, Tournay E, Hulin C, Chretien ML, Royer B, et al. Development and validation of a cytogenetic prognostic index predicting survival in multiple myeloma. J Clin Oncol. 2019;37:1657–65. doi: 10.1200/JCO.18.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies F, et al. A high-risk, double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33:159–70. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinhold N, Salwender HJ, Cairns DA, Raab MS, Waldron G, Blau IW, et al. Chromosome 1q21 abnormalities refine outcome prediction in patients with multiple myeloma - a meta-analysis of 2,596 trial patients. Haematologica. 2021;106:2754–8. doi: 10.3324/haematol.2021.278888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaughnessy JD, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. Avalidated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 23.Sawyer JR, Tian E, Walker BA, Wardell C, Lukacs JL, Sammartino G,, et al. An acquired high-risk chromosome instability phenotype in multiple myeloma: Jumping 1q Syndrome. Blood. Cancer J. 2019;9:62. doi: 10.1038/s41408-019-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gay F, Musto P, Rota-Scalabrini D, Bertamini L, Belotti A, Galli M, et al. Carfilzomib with cyclophosphamide and dexamethasone or lenalidomide and dexamethasone plus autologous transplantation or carfilzomib plus lenalidomide and dexamethasone, followed by maintenance with carfilzomib plus lenalidomide or lenalidomide alone for patients with newly diagnosed multiple myeloma (FORTE): a randomised, open-label, phase 2 trial. Lancet Oncol. 2021;22:1705–20. doi: 10.1016/S1470-2045(21)00535-0. [DOI] [PubMed] [Google Scholar]
- 25.Oliva S, De Paoli L, Ruggeri M, Caltagirone S, Troia R, Oddolo D, et al. A longitudinal analysis of chromosomal abnormalities in disease progression from MGUS/SMM to newly diagnosed and relapsed multiple myeloma. Ann Hematol. 2021;100:437–43. doi: 10.1007/s00277-020-04384-w. [DOI] [PubMed] [Google Scholar]
- 26.Chng WJ, Dispenzieri A, Chim CS, Fonseca R, Goldschmidt H, Lentzsch S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–77. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 27.Ross FM, Avet-Loiseau H, Ameye G, Gutierrez NC, Liebisch P, O’Connor S, et al. Report from the European Myeloma Network on interphase FISH in multiple myeloma and related disorders. Haematologica. 2012;97:1272–7. doi: 10.3324/haematol.2011.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Jia S, Chu Y, Tian B, Gao Y, Zhang C, et al. Chromosome 1q21 gain is an adverse prognostic factor for newly diagnosed multiple myeloma patients treated with bortezomib-based regimens. Front Oncol. 2022;12:938550. doi: 10.3389/fonc.2022.938550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Agostino M, Zaccaria GM, Ziccheddu B, Rustad EH, Genuardi E, Capra A, et al. Early relapse risk in patients with newly diagnosed multiple myeloma characterized by next-generation sequencing. Clin Cancer Res. 2020;26:4832–41. doi: 10.1158/1078-0432.CCR-20-0951. [DOI] [PubMed] [Google Scholar]
- 30.Foltz SM, Gao Q, Yoon CJ, Sun H, Yao L, Li Y, et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020;11:2666. doi: 10.1038/s41467-020-16434-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertamini L, Oliva S, Rota-Scalabrini D, Paris L, Morè S, Corradini P, et al. High Levels of circulating tumor plasma cells as a key hallmark of aggressive disease in transplant-eligible patients with newly diagnosed multiple myeloma. J Clin Oncol. 2022;40:3120–31. doi: 10.1200/JCO.21.01393. [DOI] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijhof IS, Casneuf T, van Velzen J, van Kessel B, Axel AE, Syed K, et al. CD38 expression and complement inhibitors affect response and resistance to daratumumab therapy in myeloma. Blood. 2016;128:959–70. doi: 10.1182/blood-2016-03-703439. [DOI] [PubMed] [Google Scholar]
- 34.Smith EL, Harrington K, Staehr M, Masakayan R, Jones J, Long TJ, et al. GPRC5D is a target for the immunotherapy of multiple myeloma with rationally designed CAR T cells. Sci Transl Med. 2019;11:eaau7746. doi: 10.1126/scitranslmed.aau7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel A, Smith AC, Masih-Khan E, Samuel P, Lajkosz K, Bhella S, et al. Chromosome 1q abnormalities are not an independent predictor of survival in multiple myeloma. Blood. 2022;140:4354–5. doi: 10.1182/blood-2022-167113. [DOI] [Google Scholar]
- 36.Lannes R, Samur M, Perrot A, Mazzotti C, Divoux M, Cazaubiel T, et al. In multiple myeloma, high-risk secondary genetic events observed at relapse are present from diagnosis in tiny, undetectable subclonal populations. J Clin Oncol. 2023;41:1695–702. doi: 10.1200/JCO.21.01987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapoor P, Schmidt T, Jacobus S, Wei Z, Fonseca R, Callander NS, et al. OAB-052: Impact of chromosome 1 abnormalities on newly diagnosed multiple myeloma treated with proteasome inhibitor, immunomodulatory drug, and dexamethasone: analysis from the ENDURANCE ECOG-ACRIN E1A11 trial. Clin Lymphoma Myeloma Leuk. 2021;21:S33–4. doi: 10.1016/S2152-2650(21)02126-1. [DOI] [Google Scholar]
- 38.Ziccheddu B, Da Vià MC, Lionetti M, Maeda A, Morlupi S, Dugo M, et al. Functional impact of genomic complexity on the transcriptome of multiple myeloma. Clin Cancer Res. 2021;27:6479–90. doi: 10.1158/1078-0432.CCR-20-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chari A, Minnema MC, Berdeja JG, Oriol A, van de Donk NWCJ, Rodríguez-Otero P, et al. Talquetamab, a T-cell—redirecting GPRC5D bispecific antibody for multiple myeloma. N Engl J Med. 2022;387:2232–44. doi: 10.1056/NEJMoa2204591. [DOI] [PubMed] [Google Scholar]
- 40.Mailankody S, Devlin SM, Landa J, Nath K, Diamonte C, Carstens EJ, et al. GPRC5D-targeted CAR T cells for myeloma. N Engl J Med. 2022;387:1196–206. doi: 10.1056/NEJMoa2209900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiedemann R, Mahdipour-Shirayeh A, Erdmann N, Tagoug I. OAB-010: Gain(1q) promotes mitochondrial oxidative phosphorylation and suppresses interferon response and tumor immunity in multiple myeloma and other human cancers. Clin Lymphoma Myeloma Leuk. 2021;21:S7. doi: 10.1016/S2152-2650(21)02084-X. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
After the publication of this article, data collected for this analysis and related documents will be made available to others upon reasonably justified request, which needs to be written and addressed to the attention of the corresponding author FG at the following e-mail address: francesca.gay@unito.it. The sponsor of the UNITO-MM-01/FORTE trial, the University of Torino (Italy), via the corresponding author FG, is responsible for evaluating and eventually accepting or refusing every request to disclose data and their related documents, in compliance with the ethical approval conditions, in compliance with applicable laws and regulations, and in conformance with the agreements in place with the involved subjects, the participating institutions, and all the other parties directly or indirectly involved in the participation, conduct, development, management and evaluation of this analysis.