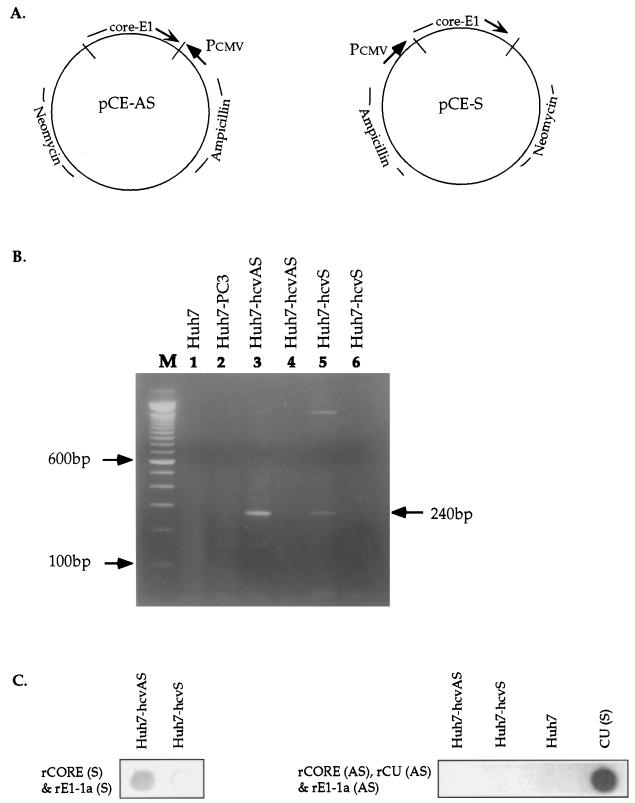
FIG. 2.
Generation and analysis of human cell lines expressing HCV genomic and antigenomic RNAs. (A) Construction of expression plasmids pCE-AS and pCE-S. The entire core and E1 genes of HCV genotype 1a were aligned in either the sense orientation (pCE-S) or the antisense orientation (pCE-AS) relative to the human cytomegalovirus major immediate-early promoter regulatory region (Pcmv). Huh7 cells were transfected with plasmid pCE-S or pCE-AS, and stable transformants, designated Huh7-hcvS and Huh7-hcvAS, were selected as described in Materials and Methods. (B) Analysis of HCV RNA expression in cell lines Huh7-hcvS and Huh7-hcvAS by RT-PCR. Total RNA was extracted from cells, digested with DNase I prior to reverse transcription in the presence of a mixture of hexamer (pdN6) oligonucleotides (lanes 3 and 5), and then amplified with primers c256 and c186 (30). The arrow on the right denotes the appropriate expected band of 240 bp. In lanes 4 and 6, the reverse transcription step was eliminated prior to PCR amplification under conditions identical to those used for lanes 3 and 5, respectively. In lanes 1 and 2, RNA from negative control cell lines Huh7 and Huh7-PC3 was analyzed by the same method as in lanes 3 and 5. Lane M contains size markers. (C) Northern dot blot analysis of RNA from transfected or control cell lines with strand-specific HCV riboprobes. CU (S) designates a genomic-strand synthetic HCV RNA which was derived from the pCU clone and which served as a positive control for the antisense riboprobes. See the legend to Fig. 1 for an explanation of other designations.