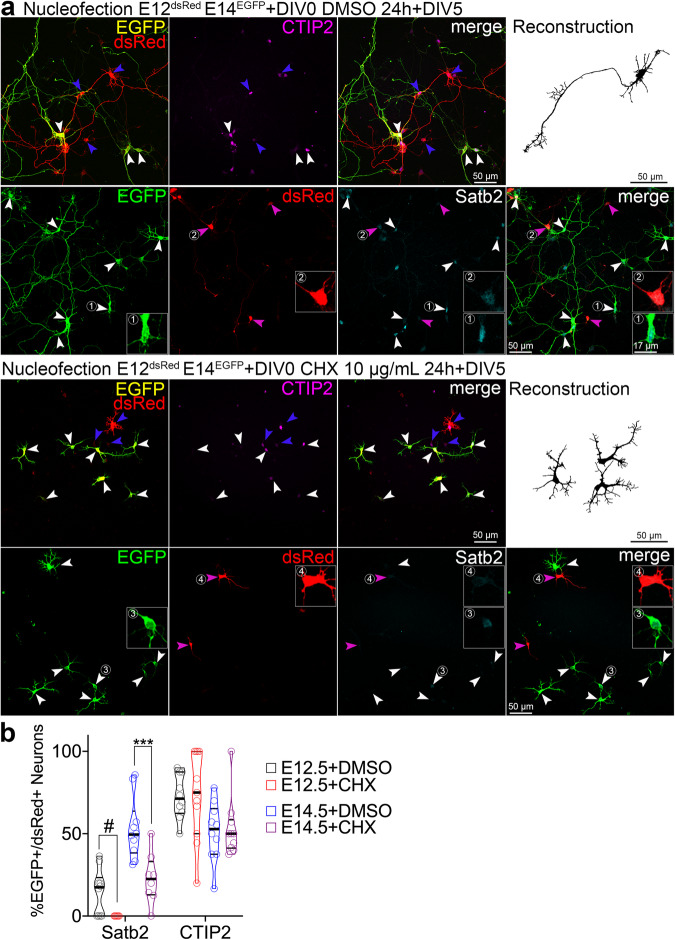
Fig. 1. Neuronal Satb2 expression requires a critical window of protein translation in precursor cells.
a Images of immunolabeled primary cells from E12.5 embryos nucleofected to express dsRed, and from E14.5 embryos to express EGFP, mixed and plated on a single glass coverslip. Two hours post-plating, cells were treated with vehicle (DMSO) or cycloheximide (CHX) for 20 hours, followed by medium change, and fixed at day-in-vitro 5 (DIV5). Upper panels show staining using rat anti-CTIP2, goat anti-EGFP, and rabbit anti-RFP, the latter one recognizing both EGFP and dsRed. For this reason, E12.5 cells in this case were recognized as solely expressing dsRed (blue arrowheads), but the E14.5 ones, both EGFP and dsRed (white arrowheads). Lower panels show anti-Satb2, anti-EGFP and anti-dsRed immunostaining with no cross-reacting antibodies; in this case, E12.5-derived cells express dsRed (blue arrowheads) and E14.5 ones EGFP (white arrowheads), as expected. Representative neuronal morphology is demonstrated as a semi-automated, EGFP- or dsRed-based reconstruction. (1-2) Example E14.5 (1,3) or E12.5 (2,4) cortex-derived cells immunolabeled with an antibody anti-Satb2. b Quantification of the cell identity markers in DIV5 neurons derived from E12.5 or E14.5 cortex. Lines on violin plot indicate median and quartiles. For statistics, Satb2, one-way ANOVA with Bonferroni’s multiple comparisons test; E14.5 + DMSO vs. E14.5 + CHX, p = 0.0007; CTIP2, Kruskal-Wallis test with Dunn’s multiple comparisons test. # indicates a comparison between fractions of Satb2-positive E12.5 DMSO- and E12.5 CHX-treated group, the latter represented by no positive cells. Data were collected from three independent cultures. *** p < 0.001. Refer to Supplementary Data S1 for detailed information on numerical values.