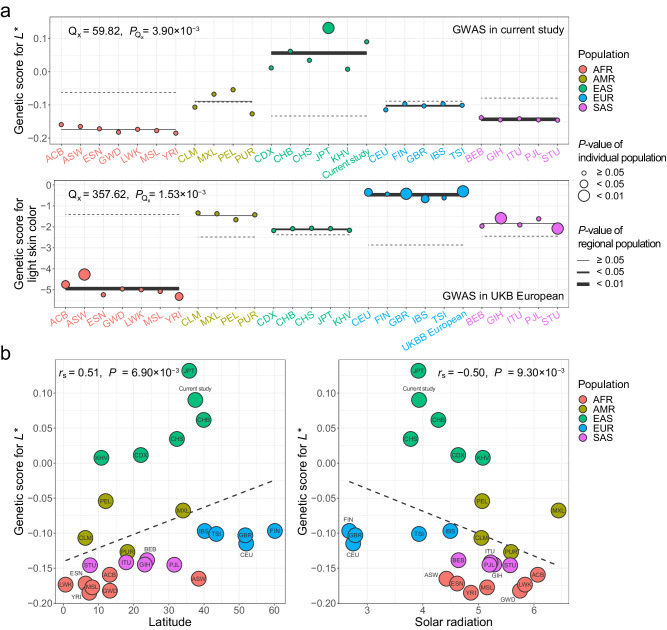
Fig. 4. Signals of polygenic adaptation for L* across the 1000 Genomes Project phase 3 populations.
a Distribution of the estimated genetic score for L* across the 1000 Genomes Project populations and results for polygenic adaptation based on the current GWAS (top) and the UK Biobank European GWAS (bottom). A test statistic for overdispersion of genetic scores (Qx) and P-values are presented at the top of each plot (two-sided). b Estimated genetic scores for L* based on the current GWAS are plotted against environmental factors: the absolute latitude of each population (left) and annual solar radiation (right). The regression lines (dashed lines) show the linearity between the genetic score (y-axis) and environmental factors (x-axis). Spearman’s correlation (rs) and P-values are presented at the top of each plot. The P-value of Spearman’s correlation coefficient was estimated using a two-sided test under the null distribution of all possible permutations. Abbreviations: AFR African, AMR admixed American, EAS East Asian, EUR European, SAS South Asian, ACB African Caribbean in Barbados, ASW African Ancestry in Southwest USA, ESN Esan in Nigeria, GWD Gambian in Western Division, Mandinka, LWK Luhya in Webuye, Kenya, MSL Mende in Sierra Leone, YRI Yoruba in Ibadan, Nigeria, CLM Colombian in Medellín, Colombia, MXL Mexican Ancestry in Los Angeles, CA, USA, PEL Peruvian in Lima, Peru, PUR Puerto Rican in Puerto Rico, CDX Chinese Dai in Xishuangbanna, China, CHB Han Chinese in Beijing, China, CHS Southern Han Chinese, China, JPT Japanese in Tokyo, Japan, KHV Kinh in Ho Chi Minh City, Vietnam, KOR Korean in the current study, CEU Utah residents with ancestry from Northern and Western Europe, FIN Finnish in Finland, GBR, British from England and Scotland, IBS Iberian Populations in Spain, TSI Toscani in Italy, UKBB European in the UK Biobank, BEB Bengali in Bangladesh, GIH Gujarati Indians in Houston, Texas, USA, ITU Indian Telugu in the UK, PJL Punjabi in Lahore, Pakistan, STU Sri Lankan Tamil in the UK.