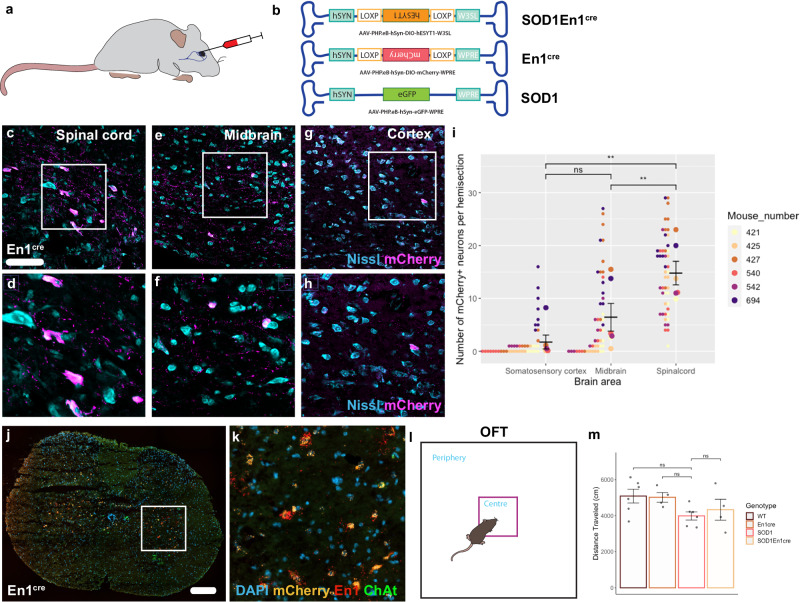
Fig. 7. Characterization of systemic overexpression of ESYT1 obtained by an AAV-PHP.eB vector.
a Cartoon depicting systemic administration via retroorbital intravenous injections of the viral constructs. b AAV vectors forcing overexpression of ESYT1, mCherry, or eGfp were injected in SOD1G93A;En1cre, En1cre, and SOD1G93A mice respectively at a volume of 100 μl per mouse. c–h Microphotograph of spinal cord (c–d), midbrain (e–f), and cortex (g–h) coronal sections of En1cre mice showing cre-dependent mCherry overexpression upon AAV-PHP.eB-hSYN-DIO-mCherry-W3SL administration. mCherry protein investigation shows successful viral targeting of the lumbar segments of the spinal cord and significantly higher transfection of the spinal cord compared to midbrain and cortex (Nissl in light blue, mCherry in magenta). Higher magnification images are shown in (d, f, h). i Quantification of mCherry positive neurons, larger dots represent the mean per mouse, while smaller dots represent the number of positive neurons per hemi section (Repeated measures ANOVA and Tukey’s post hoc: SC vs midbrain P = 0.001, SC vs cortex P = 0.0021, midbrain vs cortex P = 0.149, n = 8 per mouse and N = 6, two-tailed). j–k Microphotograph of the mCherry overexpression in the ventral horns of the lumbar spinal cord of an En1cre mouse upon systemic administration of the viral vector, using RNAscope. mCherry expression is specific to En1 positive neurons, thus selective for V1 interneurons (DAPI in blue, mCherry in orange, En1 in red, Chat in green). l Schematic depiction of the open field test (OFT), mice were assessed for general locomotor activity with the aid of automated tracking. m OFT revealed no significant differences in the distance traveled in any of the experimental conditions suggesting no deleterious effect after systemic delivery (One-way ANOVA and Dunnett’s post hoc: WT P = 0.0841, En1cre P = 0.1674, SOD1G93A;En1cre P = 0.8586, WT N = 6, En1cre N = 4, SOD1G93A N = 6, SOD1G93A;En1cre N = 4). Scale bar in c = 100 μm and in j = 200 μm. All graphs show mean values ± SEM. Source data are provided as a Source Data file.