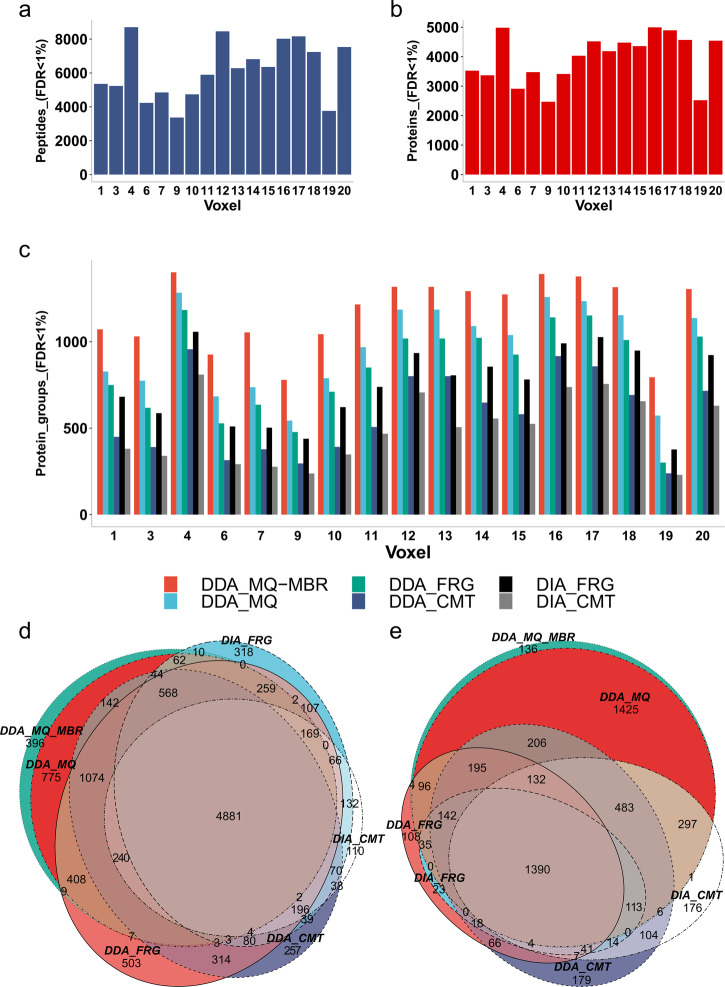
Fig. 2. Qualitative analysis of peptides and proteins identified across FFPE tissue voxels.
a Total number of unique peptides identified using a multi-search engine strategy (MaxQuant – “MQ”, MSFragger – “FRG”, Comet – “CMT”) applied to both DDA and DIA data across 17 BFPT voxels. b Total number of proteins identified using a multisearch engine strategy (MaxQuant, MSFragger, Comet) applied to both DDA and DIA data across 17 BFPT voxels. c Comparison of MaxQuant with matches between runs (DDA_MQ-MBR), MaxQuant (without MBR) (DDA_MQ), MSFragger and Comet search engines applied to both DDA (DDA_FRG and DDA_CMT respectively) and DIA data (DIA_FRG and DIA_CMT respectively) plotted as number of protein groups identified. The data suggest that MaxQuant with MBR function performs better than other search engines. In addition, c shows that DDA data yields more identified protein groups compared to DIA data regardless of the search engine used. d The Venn diagram shows the overlap of peptides identified in MaxQuant with MBR, MaxQuant (without MBR), MSFragger and Comet. The Venn diagram shows that MaxQuant with MBR function identifies the most peptides while most peptides identified in other search engines are included in the MaxQuant MBR search result. e MaxQuant with MBR function identifies most proteins compared to MSFragger and Comet, regardless of the data used. Figure e is consistent with d, which describes the same result at the peptide level.