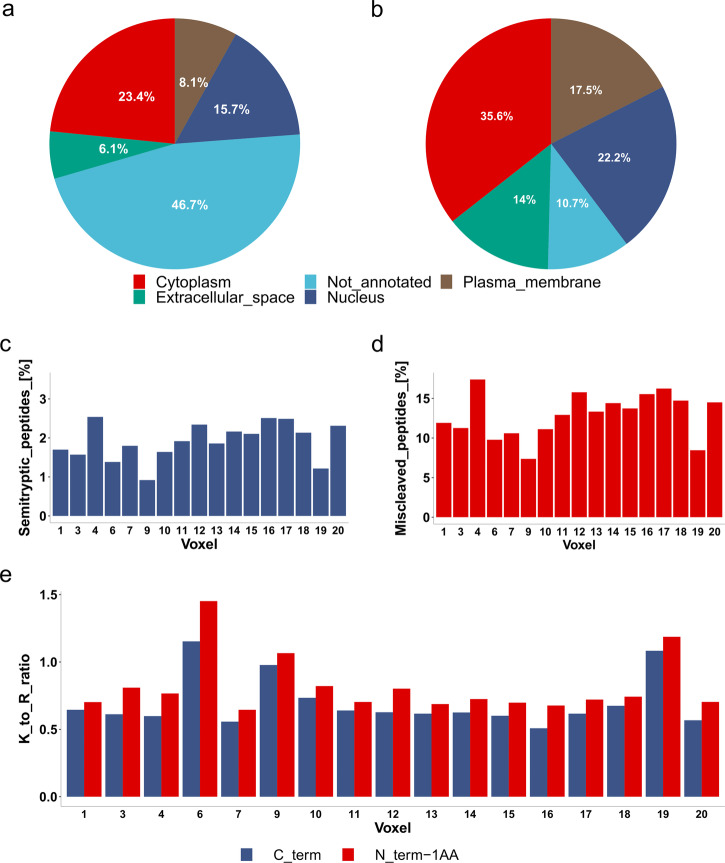
Fig. 3. Subcellular protein localization and trypsin digestion miscleavage.
a Protein subcellular localization retrieved from Uniprot Subcellular Localization. The percentage of proteins in a given localization was determined using keyword analysis. The considered subcellular localizations in the analysis were cytoplasm (red), extracellular space (green), nucleus (dark blue), and plasma membrane (brown). b Protein subcellular localization from Gene Ontology (GO). The percentage of proteins in a particular localization was calculated using the same method as in a. c The percentage of total semi-tryptic peptides across 17 voxels. d The percentage of total miscleaved peptides across 17 voxels. e The ratio of trypsin cleavage at lysine and arginine residues. The trypsin preference of lysine over arginine at the C-term of identified tryptic peptides shown in blue or at the nearest upstream tryptic peptide (identified peptide N_term minus one aminoacid) shown in red was determined. The analysis shows an overall preference for tryptic cleavage at arginine in almost all voxels.