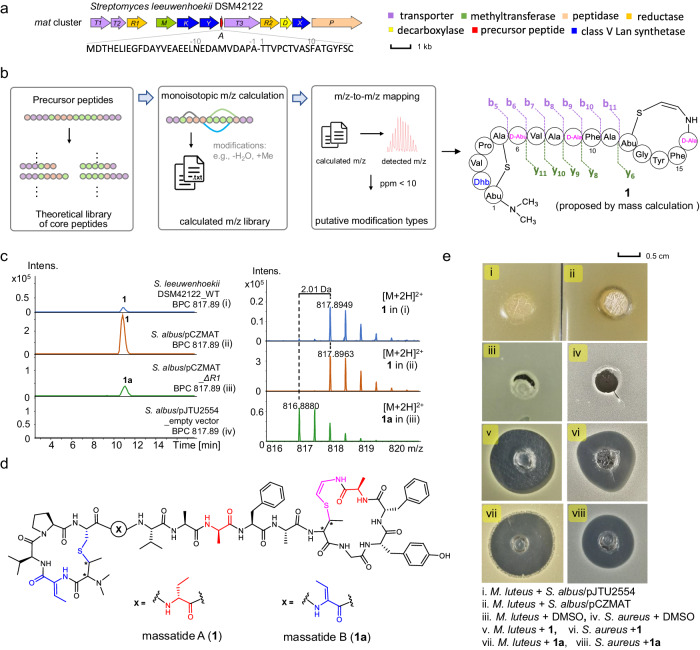
Fig. 3. Discovery and BGC characterization of massatides.
a The mat BGC and amino acid sequence of the precursor MatA. b Mass mapping pipeline and proposed structure of 1 based on tandem mass analysis. Calculation details are in the Supplementary note. c UPLC-HRMS analysis of Streptomyces leeuwenhoekii DSM42122 wild-type strain (i), S. albus/pCZMAT (ii, heterologous expression of mat BGC), S. albus/pCZMAT_ΔR1 (iii, matR1 deletion) and S. albus/pJTU2554 (iv, empty vector). d The structure of massatide. AviCys motifs are shown in pink. D-amino acids are shown in red. Other noncanonical amino acids are shown in blue. e Bioactivity of massatides against S. aureus and M. luteus (Fermentation culture was used in i and ii, and pure compounds or DMSO were used in iii–viii). For raw data, see Figure S3. Source data are provided in MassIVE (10.25345/C5PC2TM3Q).