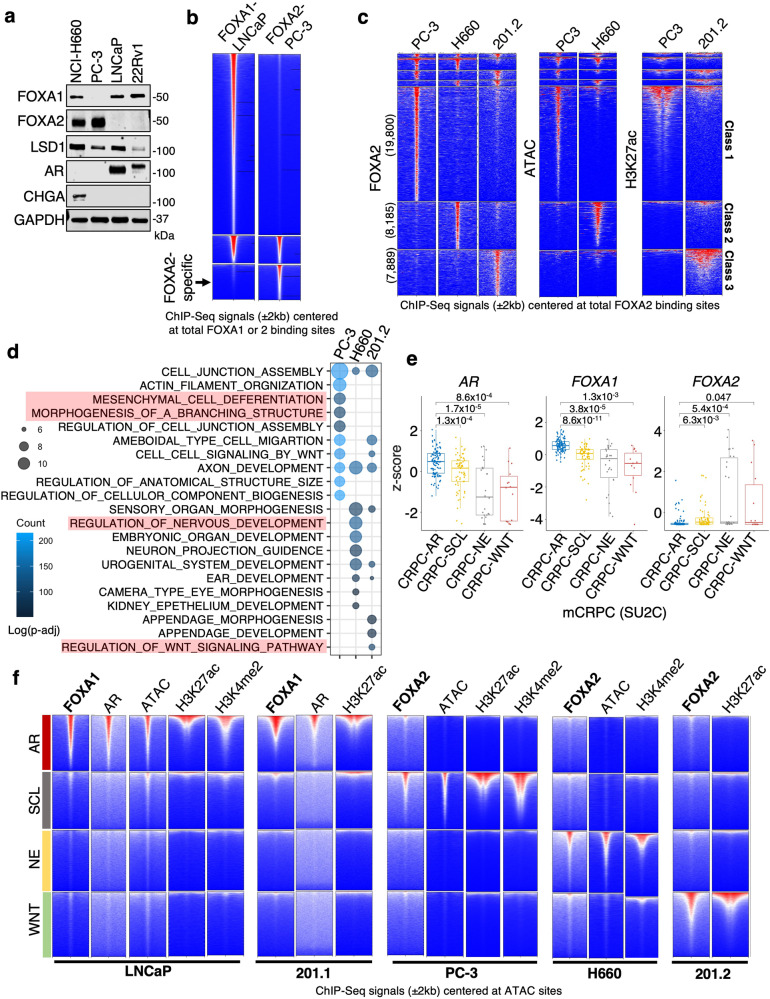
Fig. 2. FOXA2 binds to distinct classes of enhancers in three molecular subtypes of AR-independent CRPC.
a Immunoblotting for indicated proteins in PCa cell lines (n = 3 independent experiments). b Heatmap view for the ChIP-FOXA1 or FOXA2 centered at the FOXA1 and FOXA2 sites in LNCaP or PC-3 cells. c Heatmap view for the peaks of FOXA2, ATAC, or H3K27ac in PC-3, NCI-H660, or 201.2 models centered at the FOXA2 sites. d GO annotation was performed on genes associated with FOXA2 peaks that are unique to PC-3 (class 1), NCI-H660 (class 2), and 201.2 (class 3) models. e Boxplot of mRNA expression (z-score) of AR, FOXA1, and FOXA2 in previously defined CRPC subtypes—AR, SCL, NE, and WNT, using SU2C mCRPC cohort (CRPC-AR, n = 104; CRPC-SCL, n = 62; CRPC-NE, n = 26; CRPC-WNT, n = 14; center: median; box: 25th–75th IQR; whiskers: 1.5x IQR; outliers: individual data points; statistical significance determined by unpaired two-sided t-test). f Heatmap view for the ChIP-seq signal of indicated proteins centered at specific chromatin sites exhibiting different ATAC signatures for CRPC subtypes. ns (P > 0.05), *(0.01 < P < 0.05), **(0.001 < P < 0.01), ***(P < 0.001), and ****(P < 0.0001) were used to indicate the levels of P-value.