Abstract
Glioblastoma is the most common cancer in the brain, resistant to conventional therapy and prone to recurrence. Therefore, it is crucial to explore novel therapeutics strategies for the treatment and prognosis of GBM. In this study, through analyzing online datasets, we elucidated the expression and prognostic value of POLR2J and its co-expressed genes in GBM patients. Functional experiments, including assays for cell apoptosis and cell migration, were used to explore the effects of POLR2J and vorinostat on the proliferation and migration of GBM cells. The highest overexpression of POLR2J, among all cancer types, was observed in GBM. Furthermore, high expression of POLR2J or its co-expressed genes predicted a poor outcome in GBM patients. DNA replication pathways were significantly enriched in the GBM clinical samples with high POLR2J expression, and POLR2J suppression inhibited proliferation and triggered cell cycle G1/S phase arrest in GBM cells. Moreover, POLR2J silencing activated the unfolded protein response (UPR) and significantly enhanced the anti-GBM activity of vorinostat by suppressing cell proliferation and inducing apoptosis. Additionally, POLR2J could interact with STAT3 to promote the metastatic potential of GBM cells. Our study identifies POLR2J as a novel oncogene in GBM progression and provides a promising strategy for the chemotherapeutic treatment of GBM.
Keywords: Glioblastoma, POLR2J, proliferation, cell migration, prognosis
Introduction
Glioma is one of the most prevalent types of primary malignant brain tumors in adults, accounting for approximately 81% of malignant brain tumors, with a 5-year overall survival rate of less than 10% [1,2]. The standard treatment for GBM patients includes surgical resection followed by radiotherapy with concurrent and adjuvant temozolomide chemotherapy, yet prognosis remains poor [3]. Given the genetic instability, highly invasiveness, angiogenesis, resistance to conventional therapy, and proneness to recurrence [4], it is crucial to identify novel targets for the treatment and prognosis of GBM.
In eukaryotes, RNA polymerase II is a multiprotein complex responsible for transcribing DNA into mRNA precursors, as well as most small nuclear RNA and microRNA [5]. RNA polymerase II is composed of nine subunits, including RPB1, RPB2, RPB3, RPB4, RPB5, RPB6, RPB7, RPB8 and RPB11, includes DNA-directed RNA polymerase II subunit J-1 (POLR2J, also known as RPB11), which is integral to its function [6,7]. Furthermore, RNA polymerase II assembles with general transcription factors to form a pre-initiation complex which opens promoter DNA to initiate transcription [8]. Notably, transcription factor ATF4, a vital component of RNA polymerase II containing POLR2J, activates transcription by directly interacting with the RNA polymerase II at the heterodimer region of α-like subunits (RPB3-RPB11) [9]. Meanwhile, RPB3, another crucial subunit of RNA polymerase II, enhances ATF4 activation through promoter recognition [10]. Interestingly, POLR2J has been identified as a promising prognostic biomarker for patients with testicular germ cell tumor [11] and is overexpressed in rectal tumor organoids [12]. However, the mechanism responsible for the dynamic properties of POLR2J in tumor progression have yet to be fully elucidated.
In this study, we explored the fundamental functions and mechanisms of POLR2J in GBM. Firstly, POLR2J exhibits maximal upregulation in GBM compared to normal tissues across all cancer types. Furthermore, POLR2J promotes cell proliferation, metastasis, and epithelial-mesenchymal transition (EMT) in GBM. In addition, POLR2J suppression enhanced the anti-GBM activity of vorinostat. Collectively, these findings suggest that POLR2J could serve as a potential prognostic biomarker and therapeutic target for GBM.
Materials and methods
Data collection
The UALCAN portal (http://ualcan.path.uab.edu) was used to evaluate the mRNA and protein expression level of POLR2J. The potential value of POLR2J and its co-expression genes in GBM prognosis were analyzed via OS from the SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp) and ExSurv database (https://exsurv.soic.iupui.edu). Gene co-expression analysis and Gene Set Enrichment Analysis (GSEA) of POLR2J were performed utilizing LinkedOmics (http://www.linkedomics.org/admin.php). The “HiSeq RNA” platform and “TCGA_GBM” cohort were selected for the analysis. The correlation between POLR2J and STAT3 expression was explored using the cBioPortal (http://www.cbioportal.org).
Reagents and antibodies
Minimum Essential Medium (MEM) and High-glucose Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Hyclone (Logan, UT, USA) for cell culture. Trypsin-EDTA Solution (0.25%) was purchased from Gibco (Grand Island, NY, USA). BCA Protein Assay Kit and RIPA Lysis Buffer were obtained from Beyotime Biotechnology (Shanghai, China). Pre-stained protein marker was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Jet PRIME Transfection Reagent was provided by Polyplus Transfection SA (Strasbourg, France). Vorinostat (purity >99%) was purchased from Aladdin (Shanghai, China). N-acetyl-L-cysteine (NAC) was provided by Beyotime Institute of Biotechnology (Shanghai, China). Anti-POLR2J (16403-1-AP, 1:1000 dilution), anti-E-Cadherin (20874-1-AP, 1:1000 dilution), anti-N-Cadherin (22018-1-AP, 1:1000 dilution), anti-EGFR (66455-1-Ig, 1:1000 dilution) and anti-Cyclin B1 (28603-1-AP, 1:1000 dilution) antibodies were obtained from Proteintech (Wuhan, China). The anti-p-EIF2α (Ser51) (3398, 1:1000 dilution), anti-Cyclin A2 (4656, 1:1000 dilution) and anti-Cleaved PARP (5625, 1:1000 dilution) antibodies were provided by Cell Signaling Technology (Danvers, MA, USA). The anti-STAT3 (sc-482, 1:500 dilution), anti-p-AKT1/2/3 (Ser473) (sc-7985-R, 1:500 dilution) and anti-c-Myc (sc-40, 1:500 dilution) antibodies were provided by Santa Cruz Biotechnology (Santa Cruz, CA, USA). The anti-p-STAT3 (Tyr705) (ab76315, 1:1000 dilution) antibody was obtained from Abcam (Cambridge, MA, USA). The anti-GAPDH (db106, 1:10000 dilution) antibody was obtained from Diagbio. The anti-FLAG-tag (ABT2010, 1:10000 dilution) antibody was obtained from Abbkine, Inc. (San Diego, CA, USA).
Cell culture
Glioma cell lines (T98G, U251, and A172) were obtained from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). T98G was culture with MEM containing non-essential amino acids with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. U251 and A172 cells were maintained in DMEM with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. All the cells were kept in a 37°C condition with 5% CO2 and periodically tested mycoplasma negative.
Quantitative reverse transcription-PCR (qRT-PCR)
The total RNA from cultured cell lines was extracted with Trizol reagent (Takara, Tokyo, Japan), and the concentration was measured with Nano-300 Nucleic Acid Analyzer (Allsheng, Hangzhou, China). Subsequently the total RNA was converted to cDNA by the HiScript II 1st Strand cDNA Synthesis Kit according to the manufacturers’ recommendations (Vazyme Biotech, Nanjing, China). The qRT-PCR analysis was then conducted with HiScript II Q RT SuperMix (Vazyme Biotech, Nanjing, China). The 2-ΔΔCt method was used to calculate the relative levels of target genes among groups. A list of all primer sequences is provided in Supplementary Table 1.
Plasmid and siRNA transfection
Cells were seeded into 6-well plates with a density of 1 × 105 cells per well. At 30-50% confluence, siRNA was transfected using jetPRIME transfection reagent (Polyplus Transfection SA, USA). siRNAs were obtained from GenePharma (Shanghai, China) to knockdown POLR2J and the sense sequences were shown as follows: siPOLR2J-1, 5’-AGGACACCAAGGUACCCAAUGTT-3’, siPOLR2J-2, 5’-AGAAGAAGAUCACCAUUAACATT-3’, and negative control siRNA, 5’-UUCUCCGAA CGUGUCACGUTT-3’. The POLR2J overexpressed plasmid was obtained from GenScript Biotech (Piscataway, NJ, USA).
Sulforhodamine B (SRB) assay
After 24 h siRNA transfection, U251 and A172 cells were cultured into 96-well plates at a density of 3 × 103 cells/well. At 30-50% confluence, cells were treated with 1-6 µM vorinostat at 37°C for 72 h. The following steps are performed as described previously [13]. The OD value was detected at 540 nm with a microplate reader (Bioteck, Winooski, VT, USA).
Colony formation assay
U251 and T98G cells (2 × 103/well) were seeded into 6-well plates for overnight and were then transfected with siRNA, the culture medium containing the siRNA was replaced every 2-3 days for 10-14 days at 37°C. Following the supernatant is discarded, and the 6-well plates were carefully rinsed with PBS at three times, and then fixed with 4% paraformaldehyde for 30 min and 1% crystal violet stained for 20 min at room temperature, then washed with PBS and photographed.
Cell cycle analysis
U251 cells were seeded into 6-well plates at a density of 1.2 × 105 cells/well. At 30% confluence, cells were transfected with siPOLR2J for 48 h at 37°C, and then cells were collected and fixed with cold 75% ethanol at -20°C for overnight and washed with 500 µl PBS at three times. Subsequently, 1 ml U251 cell suspensions were hatched with 5 µl Propidium iodide (PI) solution for 5 min at room temperature, and cell cycle analysis was then performed with a FACSCalibur flow cytometer (USA).
Cell apoptosis assay
U251 cells (2 × 104/well) were cultured in 6-well plates. At 30% confluence, cells were transfected with siPOLR2J for 24 h and treated with 2.5 mM NAC for 6 h, afterwards treated with 4 µM vorinostat at 37°C for 48 h. The cells were harvested, cleaned with cold PBS, and resuspended in binding buffer solution mixed with 5 µl annexin V and 5 µl PI and cultivated in the dark for 15 min according to the kit’s (BD Biosciences, USA) instructions and the fluorescence property was analyzed with a FACSCalibur flow cytometer (USA). The Flowjo software was applied to measure the figures.
Wound healing assay
The transfected cells were plated in 24-well plates for overnight. Artificial wounds were created using 10 µl pipette tip, and then rinsed with 500 µl PBS for 2-3 times to remove cell debris. Cell culture medium without FBS were added and then wound distances were recorded by microscope as 0 h distance. After 24 h, wound distance was recorded, and the percentage of wound healing was measured as (the wound distance of 0 h - 24 h)/0 h wound distance × 100%.
Transwell assay
After 12 h of serum starvation, the transfected cells were collected and resuspended in the serum-free medium with the density of 3 × 105/ml. The Transwell insert membranes were coated with or without a Matrigel for invasion or migration assay, respectively. The upper chamber (8 μm pore size) was seed with 200 µl of cell suspension and a 600 µl medium with 20% FBS was added to the lower chamber. After 24 h, cells on the bottom surface of Transwell insert membranes were fixed with methanol for 15 min and stained with 1% crystal violet for 30 min at room temperature. Images were photographed and measured by ImageJ software.
Western blotting
Cellular protein lysate buffering solution was applied for protein extraction. Treated cells were collected and dissolved with cold RIPA lysate and the protein samples were separated by using 8-15% SDS-PAGE and translocated onto a PVDF (Schleicher, USA) membrane, followed by blocked with 5% skimmed milk for 1 h at room temperature. The bands were incubated overnight with specific antibodies at 4°C. Afterwards, underwent 1 h incubation at room temperature with the corresponding secondary antibody. The bands were visualized using ECL detection system (Millipore, Germany).
Co-immunoprecipitation (Co-IP) assay
U251 cells transfected with POLR2J overexpressed plasmid for 48 h were lysed with IP buffer and centrifugation at 120,000 rpm for 30 min at 4°C. Afterwards, the cell lysates were divided equally hybridized with 20 µl mouse IgG magnetic beads and anti-Flag Immunomagnetic beads, respectively, for 16 h at 4°C with gentle shaking. Subsequently, standing on the magnetic stand for 5 min, add 500 µl of PBST (NaCl 136.89 mM, KCl 2.67 mM, Na2HPO4 8.1 mM, KH2PO4 1.76 mM, 0.5% Tween 20) to the above precipitate, and redispersed the beads by gently blowing, then flip the sample up and down for 5 min and removed the supernatant after magnetic separation and repeated the above steps 3 times. Finally, resuspend with SDS loading buffer and performed SDS-PAGE detection.
Statistical analysis
All data are presented as the mean ± SD. Statistical significance was determined by unpaired two-tailed Student’s t test or one-way ANOVA followed by Tukey’s post hoc test using GraphPad Prism 7 software. P-values <0.05 were considered statistically significant.
Results
POLR2J was overexpressed and predicted poor prognosis in GBM patients
Initially, we compared the mRNA expression of POLR2J between tumor and normal tissue across 24 cancer types using the UALCAN database. Compared with normal tissues, the most significant upregulation of POLR2J was observed in GBM, indicating the highest level of POLR2J expression among all cancer types (Figure 1A, 1B) [14,15]. Similarly, POLR2J protein levels were found to be overexpressed in GBM compared to normal tissues (Figure 1C) [16], and elevated POLR2J expression was predictive of poor outcomes of GBM patients (P=0.008, Figure 1D) [17]. Furthermore, the Kaplan-Meier plot of POLR2J in the TCGA-GBM dataset obtained from The Human Protein Atlas (https://www.proteinatlas.org/) also predicted poor prognosis of GBM patients with high expression of POLR2J (P=0.017, Figure 1E). These findings suggested that POLR2J may participate in development of glioma and that the overexpression of POLR2J was correlated with poor prognosis of glioma patients.
Figure 1.
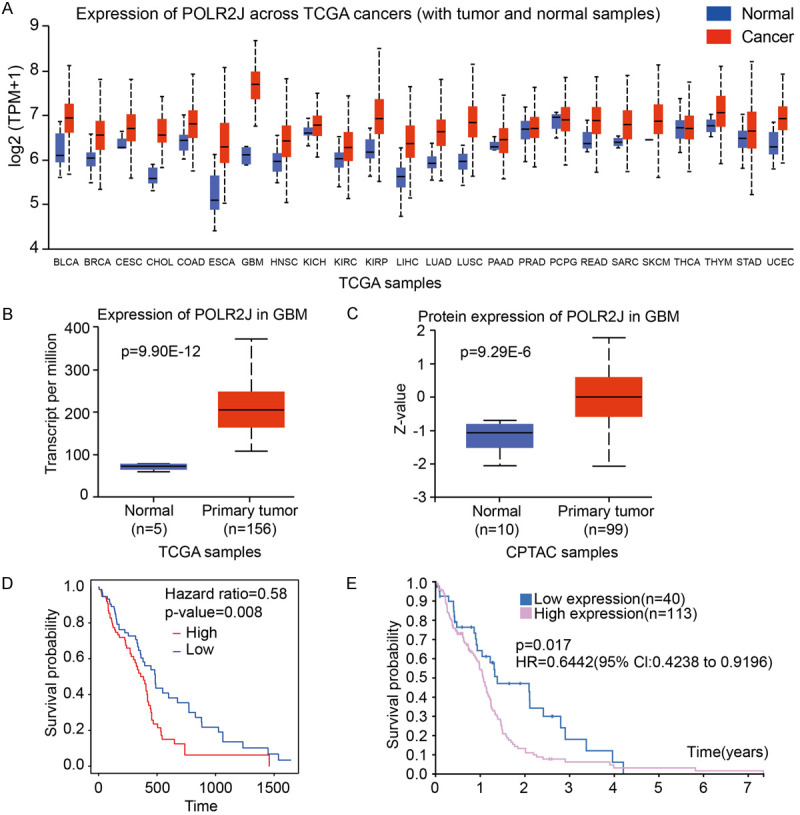
POLR2J was overexpressed and predicted poor prognosis in GBM patients. (A-C) Comparison of POLR2J expression between tumor and normal samples. Pan-cancer view (A) and mRNA Expression in Glioblastoma multiforme (B); protein expression in Glioblastoma multiforme (C). (D) Kaplan-Meier analysis of the association between POLR2J expression and OS in GBM form https://exsurv.soic.iupui.edu/. (E) The survival plot of POLR2J in TCGA-GBM samples from HPA dataset.
POLR2J co-expressed genes in GBM predicted poor prognosis
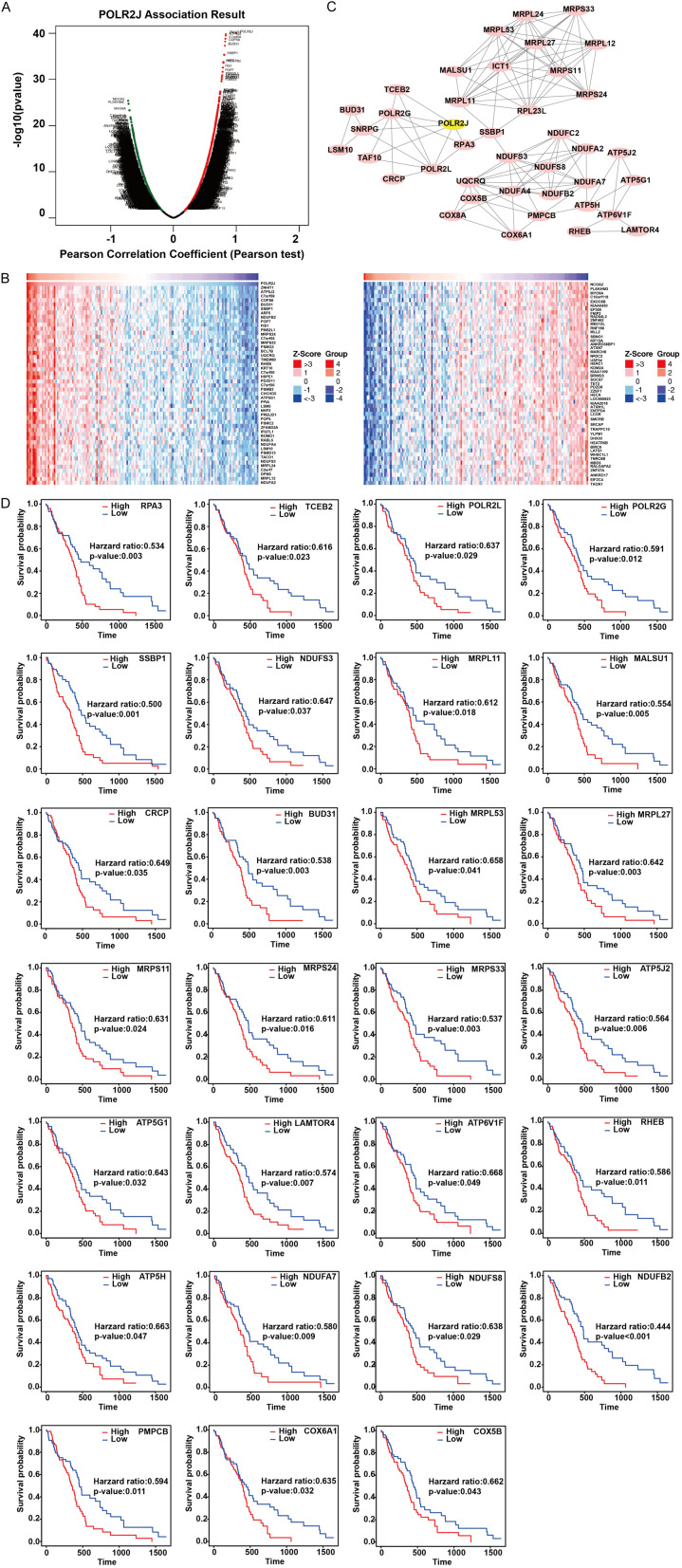
To further elucidate the prognostic value of POLR2J in GBM patients, we identified the co-expressed genes of POLR2J in GBM using LinkedOmics [18]. The Pearson coefficient test revealed 8187 positively co-expressed genes (red label) and 11,473 negatively co-expressed genes (green label) with POLR2J in GBM (Figure 2A). The top 50 crucial genes which were positively (left panel) and negatively (right panel) associated with POLR2J are shown in a heat map (Figure 2B). A network of protein-protein connections between 39 significantly co-expressed genes that had an important positive correlation with POLR2J was visualized using Cytoscape (Figure 2C) [19]. The prognostic value of these co-expressed genes was evaluated using ExSurv [17], showing that 27 of the 39 co-expressed genes were associated with poor outcomes in GBM patients (Figure 2D). This indicates that both POLR2J and its co-expressed genes are linked to worse prognoses in GBM patients.
Figure 2.
The co-expressed genes of POLR2J in GBM predicted poor prognosis. A. The whole significantly associated genes with POLR2J distinguished by Pearson test in GBM cohort. B. Heat maps of the most top 50 significant genes positively and negatively correlated with POLR2J in glioma. C. Protein-protein connection network and MCODE analysis of POLR2J co-expressed genes. D. Kaplan-Meier analysis of the association between POLR2J co-expressed genes and OS in GBM.
POLR2J suppression inhibits GBM cells proliferation
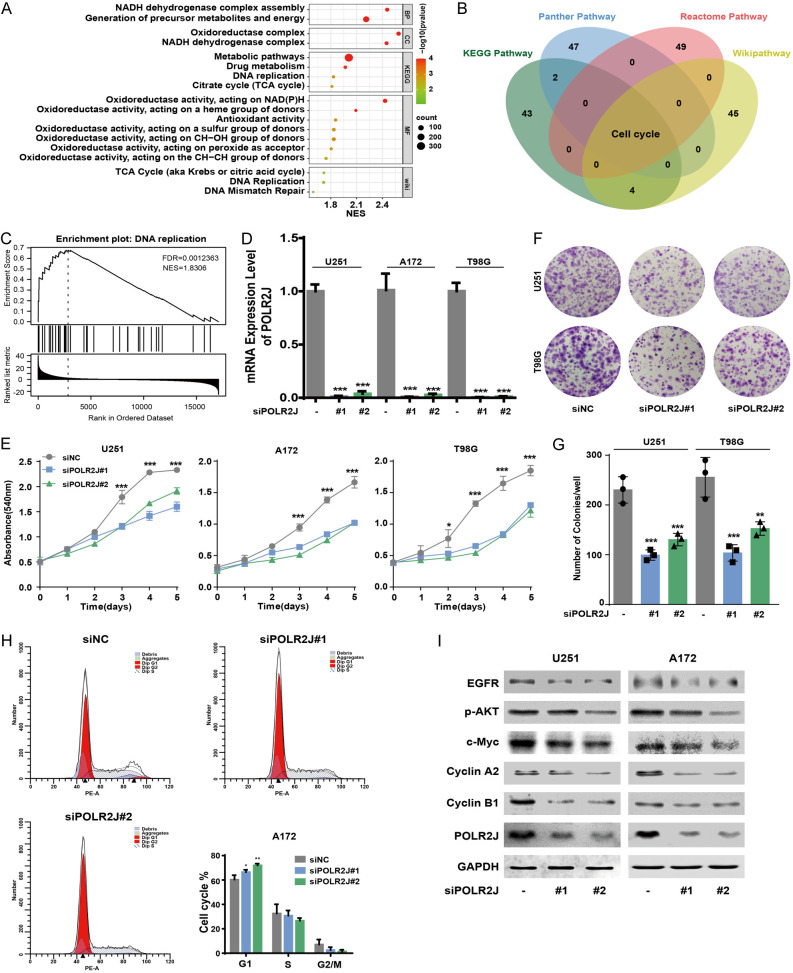
Gene Set Enrichment Analysis (GSEA) was conducted to annotate the biological function of POLR2J in GBM tumorigenesis using LinkedOmics [18]. GSEA plots demonstrated that the POLR2J signature positively correlated with pathways involved in the citrate cycle, DNA replication, mismatch repair, and metabolic processes, suggesting POLR2J may participate in the proliferation of GBM cells (Figure 3A). Additionally, a Venn diagram from over-representation analysis (ORA) generated using LinkedOmics highlighted the significant enrichment of cell cycle pathways from KEGG pathway, Panther pathway, Reactome pathway, and Wikipathway in GBM samples with high POLR2J expression (Figure 3B). GSEA further validated the positive correlation between POLR2J and DNA replication in GBM (Figure 3C) [18]. Consequently, we explored the impact of POLR2J inhibition on GBM cell proliferation, revealing substantial suppression (Figure 3D, 3E) and reduced clone formation (Figure 3F, 3G). In addition, GSEA enrichment demonstrated that POLR2J was positively correlated with cell cycle in GBM patient samples, and POLR2J silencing significantly triggered cell cycle G1/S phase arrest in GBM cells (Figure 3H). Furthermore, EGFR activation in glioblastoma promoted cellular proliferation via activation of PI3K-Akt pathways [20,21]. Moreover, POLR2J knockdown significantly inhibited EGFR and AKT pathways involved in GBM cell proliferation (Figure 3I).
Figure 3.
POLR2J suppression inhibits the proliferation of GBM cells. A. Visualization of positively functional profiles for POLR2J based on GSEA. B. The Venn diagram through over-representation analysis (ORA) using LinkedOmics demonstrated that the enrichment in the GBM clinical samples with high POLR2J expression among KEGG pathway, Panther pathway, Reactome pathway, and Wikipathway. C. GSEA was utilized to investigate the enrichment of DNA replication in GBM process according to the expression of POLR2J. D. GBM cells were transfected with siPOLR2J or negative siRNA for 48 h, and then RT-PCR was used to detect the knock-down efficiency of siPOLR2J in GBM cells. E. GBM cells were incubated with siPOLR2J or negative siRNA for 24 h, and then transferred to 96-wells plate, the cell proliferation was determined after 1, 2, 3, 4, and 5 days. F, G. GBM cells were transfected with siPOLR2J or negative siRNA, and the number of cell clone was counted. H. GBM cells were transfected with siPOLR2J or negative siRNA for 24 h, and the cell cycle distribution was tested by PI staining followed by flow cytometer. I. GBM cells were transfected with siPOLR2J or negative siRNA for 48 h, the expression of proteins was detected by western blot.
POLR2J suppression enhanced the anti-GBM activity of vorinostat
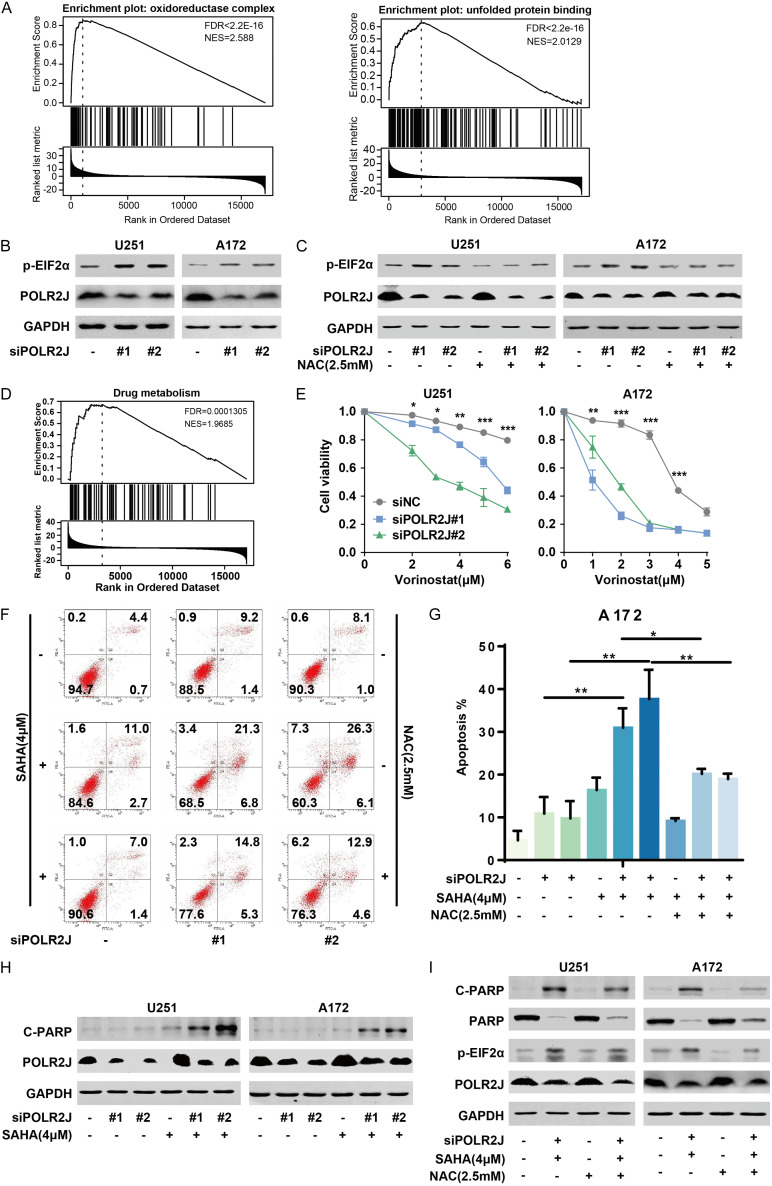
GSEA enrichment revealed that POLR2J was positively correlated with NADH dehydrogenase complex assembly, oxidoreductase complex, NADH dehydrogenase complex, and oxidoreductase activity in GBM (Figure 3A). The interplay between oxidative stress and the unfolded protein response (UPR), which triggers endoplasmic reticulum (ER) stress, is known to contribute to tumor development [22]. Furthermore, tumor cells adapt to ER stress by increasing NADPH production [23]. Therefore, we hypothesized that POLR2J might participate in cancer progression by regulating oxidoreductase activity and the UPR. GSEA, conducted using the LinkedOmics online tool, confirmed that POLR2J was positively associated with oxidoreductase activity and the unfolded protein response in GBM (Figure 4A). Our data demonstrates that POLR2J silencing significantly activated the UPR by enhancing the phosphorylation of eIF2α, a key indicator of UPR activation (Figure 4B). Additionally, N-acetylcysteine (NAC) reversed the activation of UPR induced by POLR2J silencing in GBM cells (Figure 4C). Adapting to oxidative stress has been proposed as an effective strategy to overcome chemotherapy resistance in cancer treatment [24]. Despite multimodal treatments for GBM, the prognosis remains poor, largely due to challenges in blood-brain barrier penetration and intrinsic GBM resistance. Histone deacetylase inhibitors (HDACis) have been explored as a treatment for various malignancies by altering transcriptomic profiles to induce tumor cell death. Vorinostat (SAHA) is the most advanced HDAC inhibitor to enter clinical trials for GBM. Our findings suggest that POLR2J may influence drug metabolism, and its silencing markedly enhanced the cytotoxic effect of vorinostat on GBM cells (Figure 4D and 4E). Furthermore, POLR2J knockdown increased vorinostat-induced apoptosis in GBM cells, an effect that could be mitigated by NAC treatment (Figure 4F, 4G). The cleavage of PARP, a classical marker of apoptosis, was observed when combining vorinostat with a PARP inhibitor, leading to apoptosis in GBM cells [25]. Moreover, POLR2J silencing amplified PARP cleavage induced by vorinostat (Figure 4H). This effect, as well as the expression of cleaved PARP (C-PARP) and phosphorylated eIF2α (p-eIF2α) induced by POLR2J knockdown in combination with vorinostat, was reversed by NAC (Figure 4I). This indicates that POLR2J suppression enhances the sensitivity to vorinostat, potentially through mechanisms involving PARP and eIF2α. These findings suggest that POLR2J silencing activates the UPR and strengthens the anti-GBM efficacy of vorinostat by modulating the UPR.
Figure 4.
POLR2J suppression enhanced the anti-GBM activity of vorinostat. A. GSEA was utilized to investigate the enrichment of oxidative stress and unfolded protein response in GBM according to the expression of POLR2J using LinkedOmics online tool. B. GBM cells were transfected with siPOLR2J or negative siRNA for 48 h, the expression of proteins was detected by western blot. C. GBM cells were transfected with siPOLR2J or negative siRNA for 6 h, and then cells were incubated with NAC for 48 h, finally the expression of proteins was detected by western blot. D. GSEA was utilized to investigate the enrichment of drug metabolism in GBM according to the expression of POLR2J using LinkedOmics online tool. E. GBM cells were cultured with siPOLR2J or negative siRNA for 24 h, and then incubated with vorinostat at the indicated concentrations for 72 h, SRB assay was performed to detect the proliferation of GBM cells. F, G. GBM cells were incubated with siPOLR2J or negative siRNA for 24 h, and then treated with vorinostat at the indicated concentrations for 48 h, Annexin V-FITC/PI staining assay was performed to detect the apoptosis of GBM cells. H. GBM cells were cultured with siPOLR2J or negative siRNA for 24 h, and then treated with vorinostat at the indicated concentrations for 48 h, the expression of protein was detected by western blot. I. GBM cells were transfected with siPOLR2J or negative siRNA for 24 h, pretreated with NAC for 6 h, and then incubated with vorinostat for 48 h, finally the expression of protein was detected by western blot.
POLR2J promotes the migratory and invasive abilities of GBM cells
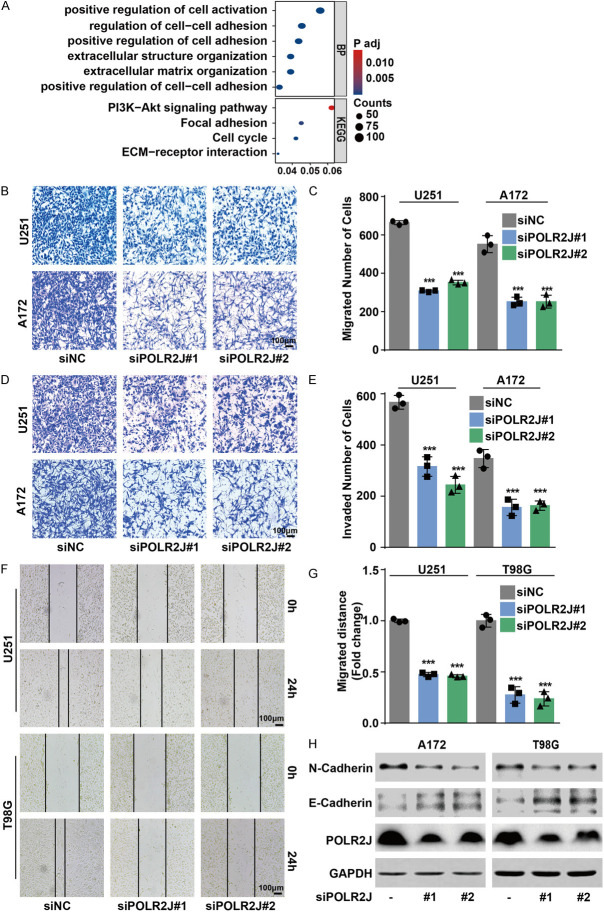
EMT is recognized as a critical process in initiating tumor invasion and metastasis, affecting cell shape, adhesion, and movement [26]. GO-KEGG analysis of genes co-expressed with POLR2J (Figure 2) similarly underscored the significance of the cell cycle pathway, as highlighted in the corresponding Figure 3B. The results also indicated the enrichment of the cell adhesion, ECM-receptor interaction pathways among genes associated with POLR2J (Figure 5A). Consequently, we posited that POLR2J plays a significant role in the EMT and metastatic potential of GBM cells. Indeed, transwell assays demonstrated that POLR2J suppression could remarkably restrict the migrative and invasive abilities of GBM cells, including U251 and A172 cells (Figure 5B-E). Likewise, wound healing assays also revealed that POLR2J inhibition could significantly inhibit the migration ability of GBM cells (Figure 5F, 5G). In addition, POLR2J knockdown suppressed the progress of EMT in GBM cells by increasing E-cadherin expression and reducing N-cadherin expression (Figure 5H). Conversely, transwell assays demonstrated that POLR2J overexpression significantly enhanced the migratory and invasive capabilities of GBM cells (Figure 6A-D). Likewise, POLR2J overexpression significantly enhanced the migrative ability of GBM cells in wound healing assays (Figure 6E, 6F). Moreover, POLR2J augmented the EMT progress in GBM cells by promoting N-cadherin expression and reducing E-cadherin expression (Figure 6G). These findings suggest that POLR2J accelerates the migratory and invasive abilities of GBM cells.
Figure 5.
POLR2J silencing inhibited the migration and invasion of glioma cells. A. GO-KEGG analysis of genes which were co-expressed with POLR2J. B, C. GBM cells were transfected with siPOLR2J or negative siRNA for 24 h, and then transferred to the upper chamber of transwell, the migrated cells were stained with crystal violet and counted. D, E. GBM cells were transfected with siPOLR2J or negative siRNA for 24 h, and then transferred to the upper chamber of transwell with a Matrigel coating on the insert membrane, the invaded cells were stained with crystal violet and counted. F, G. GBM cells were incubated with siPOLR2J or negative siRNA for 24 h and wound healing assay was performed. H. GBM cells were transfected with siPOLR2J or negative siRNA for 48 h, and then western blot was performed.
Figure 6.
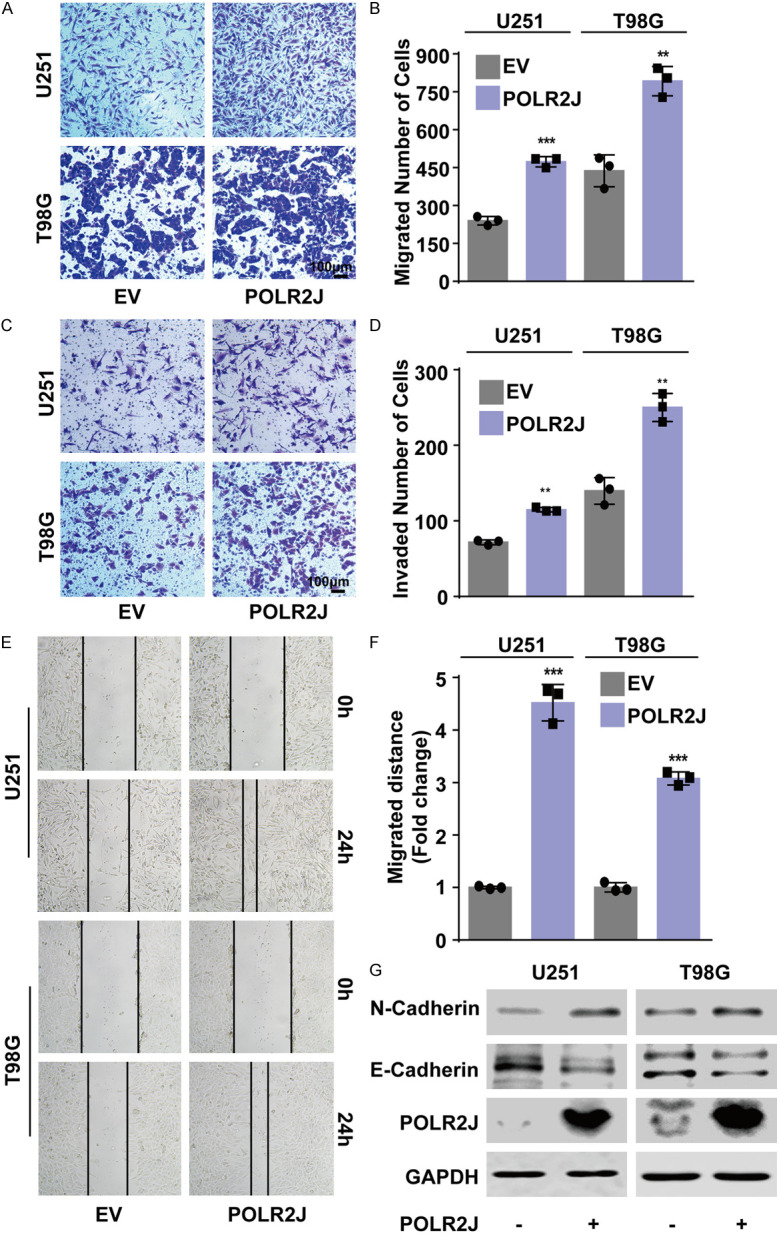
POLR2J promoted the migration and invasion of glioma cells. A, B. GBM cells were transfected with POLR2J plasmid or empty vector for 24 h, and then transferred to the upper chamber of transwell, the migrated cells were stained with crystal violet and counted. C, D. GBM cells were incubated with POLR2J plasmid or empty vector for 24 h, and then transferred to the upper chamber with a Matrigel coating on the insert membrane, the invaded cells were stained with crystal violet and counted. E, F. GBM cells were transfected with POLR2J plasmid or empty vector for 24 h, and then wound healing assay was performed. G. GBM cells were transfected with POLR2J plasmid or empty vector for 48 h, and then western blot was performed.
POLR2J interacts with STAT3
The GO-KEGG analysis presented in Figure 5A also highlighted the enrichment of PI3K-AKT and focal adhesion pathways among genes associated with POLR2J. Subsequent analysis revealed that POLR2J co-expressed with Signal Transducer and Activator of Transcription 3 (STAT3) in diffuse glioma and brain low-grade glioma patient samples (Figure 7A; diffuse glioma: Spearman =0.24, P=0.0307, Pearson =0.57, P=5.03e-8; brain low grade glioma: Spearman =0.20, P=6.58e-4, Pearson =0.12, P=0.0408) [27-30]. Given that RNA polymerase II interacts with general transcription factors to form a pre-initiation complex, thereby opening promoter DNA to initiate transcription, we hypothesized that POLR2J may interact with STAT3 to promote malignant behaviors in GBM. Interestingly, the result showed that POLR2J could bind to STAT3, which was confirmed by immunoprecipitation in U251 cells (Figure 7B). Numerous studies have demonstrated that STAT3 plays a significant role in the proliferation and metastasis of tumors [31]. To further elucidate POLR2J-regulated metastasis via the STAT3 pathway, we examined whether overexpression of POLR2J could rescue GBM migration and invasion mediated by STAT3 silencing. Wound healing assays demonstrated that POLR2J overexpression significantly enhanced the migration of GBM cells, whereas STAT3 downregulation mitigated this increased migration (Figure 7C, 7D). Similarly, a transwell assay confirmed that upregulating POLR2J expression restored GBM migration and invasion following STAT3 downregulation (Figure 7E-G). Additionally, our results indicated that the protein level of p-STAT3 Tyr705 was reduced following POLR2J knockdown (Figure 7H). Furthermore, we investigated the effects of concurrent STAT3 suppression and POLR2J overexpression. It was found that the upregulation of N-Cadherin, Cyclin A2, STAT3, and p-STAT3 Tyr705 protein levels due to POLR2J overexpression was mitigated by STAT3 inhibition (Figure 7I). These results demonstrate that POLR2J regulates metastasis and EMT in glioma cells by promoting the STAT3 signaling pathway.
Figure 7.
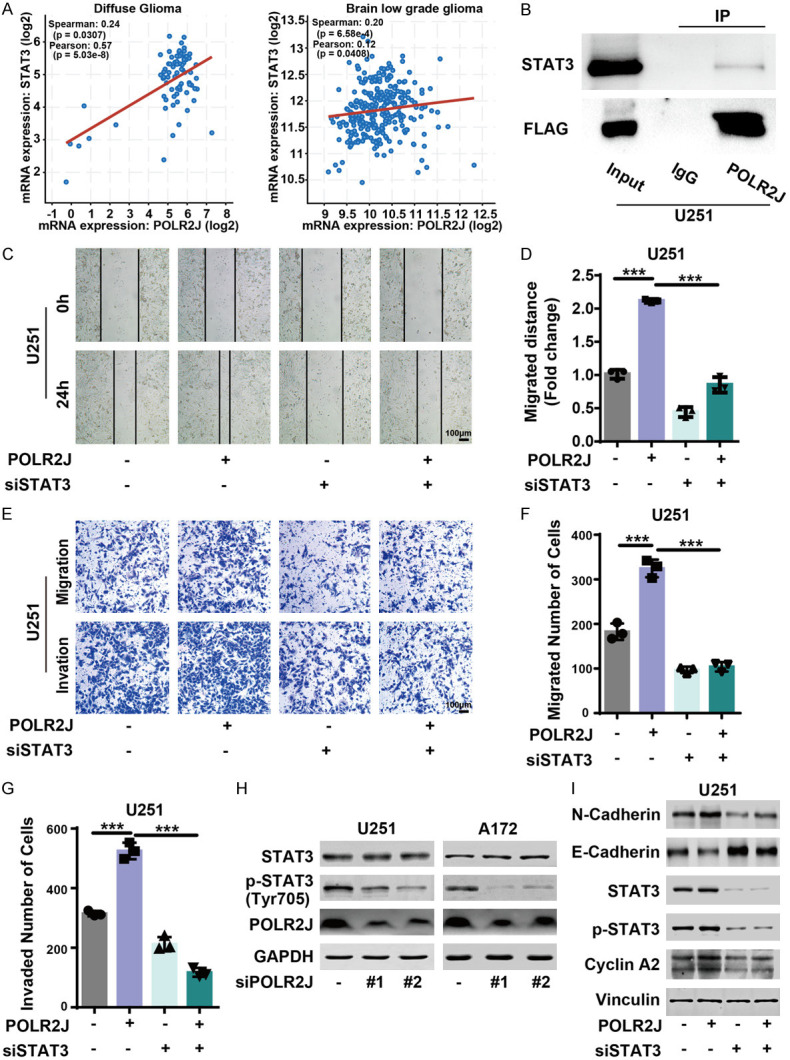
POLR2J promoted the migration of glioma cells by regulating STAT3 signalling pathway. A. The co-expression relationships between POLR2J and STAT3 were collected from cBioPortal (http://www.cbioportal.org). B. U251 cells were incubated with POLR2J plasmid for 48 h, an immunoprecipitation assay was carried out to assess the interaction between POLR2J and STAT3. C, D. GBM cells were transfected with POLR2J plasmid or empty vector for 24 h and transfected with siSTAT3 or negative siRNA for 24 h, and then wound healing assay was performed. Images were acquired under a microscope. E-G. GBM cells were transfected with POLR2J plasmid or empty vector for 24 h and transfected with siSTAT3 or negative siRNA for 24 h, and then the migration and invasion ability of GBM cells were assessed. H. GBM cells were incubated with siPOLR2J or negative siRNA for 48 h, and then western blot was performed to analyze the expression of STAT3 and p-STAT3. I. GBM cells were cultured with POLR2J plasmid or empty vector for 24 h and transfected with siSTAT3 or negative siRNA for 24 h, and then western blot was conducted to analyze the expression of proteins.
Discussion
GBM represents the most common primary malignant brain tumor, characterized by high morbidity and mortality. Due to the extensive invasion and infiltration capabilities of GBM, complete resection is challenging to achieve, and recurrence is common even after total removal [32]. Consequently, identifying promising biomarkers for the diagnosis and therapy of GBM patients is of paramount importance. This study is the first to demonstrate that POLR2J is significantly upregulated in GBM compared to normal tissues across all cancer types, with high levels of POLR2J predicting a poor outcome for GBM patients. Additionally, genes co-expressed with POLR2J were also linked to unfavorable prognoses in GBM patients. Furthermore, POLR2J suppression notably inhibited cell proliferation, and silencing POLR2J induced significant cell cycle G1/S phase arrest in GBM cells. Moreover, POLR2J reinforced the EMT progress, as well as the migratory and invasive capabilities of GBM cells. In this context, our data, for the first time, reveals that POLR2J functions as an oncogene in the tumorigenesis of GBM, presenting it as a promising prognostic and therapeutic biomarker for GBM.
Temozolomide (TMZ) is widely used in GBM treatment; however, approximately 50% of patients treated with TMZ do not respond [33]. Additionally, the overall 5-year survival rate post-TMZ treatment is only 9.8%, underscoring the urgent need for alternative GBM treatment options [34]. HDAC inhibitors, such as vorinostat, have shown tolerability as monotherapy in patients with recurrent GBM and demonstrate modest single-agent efficacy [25]. HDAC inhibitors trigger the nuclear translocation of transcription factors and activate cytoprotective autophagy, contributing to therapeutic resistance. Moreover, combining HDAC inhibitors with autophagy-modulating drugs has shown to enhance anti-cancer activity in high-risk neuroblastoma cells [35]. It has been reported that POLR2J can interact with transcription factors to activate transcription [9]. Thus, we hypothesized that silencing POLR2J might amplify the anti-cancer efficacy of vorinostat by modulating transcription factors. For the first time, this study showed that POLR2J may be involved in drug metabolism, and its silencing significantly bolstered the anti-GBM activity of vorinostat by inhibiting cell proliferation and inducing apoptosis. Endoplasmic reticulum (ER) stress/unfolded protein response (UPR) plays a critical role in GBM pathophysiology, with overexpression of the UPR marker ATF4 correlated with poor overall survival and significant in GBM progression [36]. GSEA indicated that POLR2J positively correlates with oxidoreductase activity and UPR in GBM. The UPR is mediated through three ER transmembrane protein sensors: IRE1α, PERK, and ATF6α. PERK, in response to ER stress, initiates an adaptive reaction by phosphorylating eIF2α, a commonly used marker for PERK activation [37,38]. POLR2J silencing activated the UPR by enhancing eIF2α phosphorylation, and the ROS inhibitor NAC reversed this activation in GBM cells. These findings suggest a correlation between POLR2J and UPR in GBM, where POLR2J knockdown enhances UPR activation. Given that HDAC inhibitors induce apoptosis in cancer cells through ROS accumulation [39], POLR2J silencing could potentially enhance the anti-cancer activity of vorinostat by increasing ROS accumulation, presenting a novel chemo-sensitization strategy for vorinostat treatment in GBM.
The potential mechanisms through which POLR2J promotes glioma progression remain unclear. To address this, we further explored and integrated online databases, conducting a series of in vitro experiments. Our results indicated that POLR2J is positively associated with, and can interact with STAT3. STAT3 plays a critical role in regulating cell proliferation, apoptosis, oncogenesis, and metastasis in various tumors, including glioma [40-43]. By knocking down POLR2J expression, we observed a significant reduction in the protein level of p-STAT3, suggesting that POLR2J regulates cell proliferation and invasion through STAT3 pathways. Intriguingly, our findings also revealed that suppressing STAT3 could counteract the metastasis and EMT processes mediated by POLR2J overexpression. This suggests that POLR2J promotes glioma cell metastasis by activating STAT3 signaling. Despite these findings, the mechanisms by which POLR2J interacts with STAT3 and how POLR2J specifically influences STAT3 phosphorylation require further investigation.
Conclusions
Our study has demonstrated that POLR2J is overexpressed in GBM when compared to normal tissues, and a high level of POLR2J is associated with a poor prognosis in GBM patients. Moreover, it was found that POLR2J functions as an oncogene in the tumorigenesis of GBM, and that POLR2J silencing could be a novel chemo-sensitization strategy for the use of vorinostat in treating GBM. Furthermore, POLR2J promoted the development and progression of GBM by regulating the STAT3 signaling pathway. Thus, POLR2J could serve as a novel prognostic and therapeutics biomarker for GBM treatment.
Acknowledgements
This study was supported by National Natural Science Foundation of China (82273352), Public-service Technology Research Plan of Zhejiang Province (LGF21H310002), Zhejiang Provincial Natural Science Foundation (LY19H310004, LTY21H160001, LY21H160017), Huadong Medicine Joint Funds of the Zhejiang Provincial Natural Science Foundation of China (LHDMY22H160001).
Disclosure of conflict of interest
None.
Abbreviations
- EMT
Epithelial-mesenchymal transition
- GBM
Glioblastoma multiform
- GSEA
Gene Set Enrichment Analysis
- PD-L1
Programmed cell death protein 1 ligand
- POLR2J
DNA-directed RNA polymerase II subunit J-1
- STAT3
Signal transducer and activator of transcription 3
- TICs
Tumor-infiltrating immune cells
- TMZ
Temozolomide
Supporting Information
References
- 1.Morgan LL. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2015;17:623–624. doi: 10.1093/neuonc/nou358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu W, Klockow JL, Zhang M, Lafortune F, Chang E, Jin L, Wu Y, Daldrup-Link HE. Glioblastoma multiforme (GBM): an overview of current therapies and mechanisms of resistance. Pharmacol Res. 2021;171:105780. doi: 10.1016/j.phrs.2021.105780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rong L, Li N, Zhang Z. Emerging therapies for glioblastoma: current state and future directions. J Exp Clin Cancer Res. 2022;41:142. doi: 10.1186/s13046-022-02349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevastre AS, Costachi A, Tataranu LG, Brandusa C, Artene SA, Stovicek O, Alexandru O, Danoiu S, Sfredel V, Dricu A. Glioblastoma pharmacotherapy: a multifaceted perspective of conventional and emerging treatments (Review) Exp Ther Med. 2021;22:1408. doi: 10.3892/etm.2021.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choder M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Devaux S, Lecordier L, Uzureau P, Walgraffe D, Dierick JF, Poelvoorde P, Pays E, Vanhamme L. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol Biochem Parasitol. 2006;148:60–68. doi: 10.1016/j.molbiopara.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Shpakovskii DG, Shematorova EK, Shpakovskii GV. New genes on human chromosome 7: bioinformatic analysis of a gene cluster from the POLR2J family. Bioorg Khim. 2004;30:621–625. doi: 10.1023/b:rubi.0000049773.14555.4c. [DOI] [PubMed] [Google Scholar]
- 8.Aibara S, Schilbach S, Cramer P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature. 2021;594:124–128. doi: 10.1038/s41586-021-03554-8. [DOI] [PubMed] [Google Scholar]
- 9.Proshkin SA, Shematorova EK, Shpakovski GV. The human isoform of RNA polymerase II subunit hRPB11balpha specifically interacts with transcription factor ATF4. Int J Mol Sci. 2019;21:135. doi: 10.3390/ijms21010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Angelis R, Iezzi S, Bruno T, Corbi N, Di Padova M, Floridi A, Fanciulli M, Passananti C. Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4) FEBS Lett. 2003;547:15–19. doi: 10.1016/s0014-5793(03)00659-8. [DOI] [PubMed] [Google Scholar]
- 11.Yao L, Cong R, Ji C, Zhou X, Luan J, Meng X, Song N. RNA-binding proteins play an important role in the prognosis of patients with testicular germ cell tumor. Front Genet. 2021;12:610291. doi: 10.3389/fgene.2021.610291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costales-Carrera A, Fernández-Barral A, Bustamante-Madrid P, Domínguez O, Guerra-Pastrián L, Cantero R, Del Peso L, Burgos A, Barbáchano A, Muñoz A. Comparative study of organoids from patient-derived normal and tumor colon and rectal tissue. Cancers (Basel) 2020;12:2302. doi: 10.3390/cancers12082302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Wu LW, Li ZD, Zhang MM, Wu J, Du FH, Zeng LH, Li YL. DYRK1A suppression attenuates HIF-1alpha accumulation and enhances the anti-liver cancer effects of regorafenib and sorafenib under hypoxic conditions. Int J Oncol. 2022;60:45. doi: 10.3892/ijo.2022.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, Netto GJ, Qin ZS, Kumar S, Manne U, Creighton CJ, Varambally S. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi: 10.1016/j.neo.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13:2669. doi: 10.1038/s41467-022-30342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashemikhabir S, Budak G, Janga SC. ExSurv: a web resource for prognostic analyses of exons across human cancers using clinical transcriptomes. Cancer Inform. 2016;15(Suppl 2):17–24. doi: 10.4137/CIN.S39367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarti A, Zhai G, Suzuki Y, Sarkesh S, Black PM, Muzikansky A, Loeffler JS. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J. Clin. Oncol. 2004;22:1926–1933. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- 21.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Y, Jiang M, Chen W, Zhao T, Wei Y. Cancer and ER stress: mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed Pharmacother. 2019;118:109249. doi: 10.1016/j.biopha.2019.109249. [DOI] [PubMed] [Google Scholar]
- 23.Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38:167–197. doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Avril T, Vauleon E, Chevet E. Endoplasmic reticulum stress signaling and chemotherapy resistance in solid cancers. Oncogenesis. 2017;6:e373. doi: 10.1038/oncsis.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen RD, Gajjar MK, Jensen KE, Hamerlik P. Enhanced efficacy of combined HDAC and PARP targeting in glioblastoma. Mol Oncol. 2016;10:751–763. doi: 10.1016/j.molonc.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banyard J, Bielenberg DR. The role of EMT and MET in cancer dissemination. Connect Tissue Res. 2015;56:403–413. doi: 10.3109/03008207.2015.1060970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro KD, Alpar D, Amin SB, Ashley DM, Bandopadhayay P, Barnholtz-Sloan JS, Beroukhim R, Bock C, Brastianos PK, Brat DJ, Brodbelt AR, Bruns AF, Bulsara KR, Chakrabarty A, Chakravarti A, Chuang JH, Claus EB, Cochran EJ, Connelly J, Costello JF, Finocchiaro G, Fletcher MN, French PJ, Gan HK, Gilbert MR, Gould PV, Grimmer MR, Iavarone A, Ismail A, Jenkinson MD, Khasraw M, Kim H, Kouwenhoven MCM, LaViolette PS, Li M, Lichter P, Ligon KL, Lowman AK, Malta TM, Mazor T, McDonald KL, Molinaro AM, Nam DH, Nayyar N, Ng HK, Ngan CY, Niclou SP, Niers JM, Noushmehr H, Noorbakhsh J, Ormond DR, Park CK, Poisson LM, Rabadan R, Radlwimmer B, Rao G, Reifenberger G, Sa JK, Schuster M, Shaw BL, Short SC, Smitt PAS, Sloan AE, Smits M, Suzuki H, Tabatabai G, Van Meir EG, Watts C, Weller M, Wesseling P, Westerman BA, Widhalm G, Woehrer A, Yung WKA, Zadeh G, Huse JT, De Groot JF, Stead LF, Verhaak RGW GLASS Consortium. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576:112–120. doi: 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadrkhanloo M, Entezari M, Orouei S, Ghollasi M, Fathi N, Rezaei S, Hejazi ES, Kakavand A, Saebfar H, Hashemi M, Goharrizi MASB, Salimimoghadam S, Rashidi M, Taheriazam A, Samarghandian S. STAT3-EMT axis in tumors: modulation of cancer metastasis, stemness and therapy response. Pharmacol Res. 2022;182:106311. doi: 10.1016/j.phrs.2022.106311. [DOI] [PubMed] [Google Scholar]
- 32.Uddin MS, Mamun AA, Alghamdi BS, Tewari D, Jeandet P, Sarwar MS, Ashraf GM. Epigenetics of glioblastoma multiforme: from molecular mechanisms to therapeutic approaches. Semin Cancer Biol. 2022;83:100–120. doi: 10.1016/j.semcancer.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3:198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohitesh K, Saini H, Srivastava A, Mukherjee S, Roy A, Chowdhury R. Autophagy inhibition potentiates SAHA-mediated apoptosis in glioblastoma cells by accumulation of damaged mitochondria. Oncol Rep. 2018;39:2787–2796. doi: 10.3892/or.2018.6373. [DOI] [PubMed] [Google Scholar]
- 35.Korholz K, Ridinger J, Krunic D, Najafi S, Gerloff XF, Frese K, Meder B, Peterziel H, Vega-Rubin-de-Celis S, Witt O, Oehme I. Broad-spectrum HDAC inhibitors promote autophagy through FOXO transcription factors in neuroblastoma. Cells. 2021;10:1001. doi: 10.3390/cells10051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penaranda-Fajardo NM, Meijer C, Liang Y, Dijkstra BM, Aguirre-Gamboa R, den Dunnen WFA, Kruyt FAE. ER stress and UPR activation in glioblastoma: identification of a noncanonical PERK mechanism regulating GBM stem cells through SOX2 modulation. Cell Death Dis. 2019;10:690. doi: 10.1038/s41419-019-1934-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 39.Kim HI, Seo SK, Chon SJ, Kim GH, Lee I, Yun BH. Changes in the expression of TBP-2 in response to histone deacetylase inhibitor treatment in human endometrial cells. Int J Mol Sci. 2021;22:1427. doi: 10.3390/ijms22031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, He Z, Ye J, Liu Z, She X, Gao X, Liang R. Progress in understanding the IL-6/STAT3 pathway in colorectal cancer. Onco Targets Ther. 2020;13:13023–13032. doi: 10.2147/OTT.S278013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong L, Li J, Li Q, Wang X, Medikonda R, Zhao T, Li T, Ma H, Yi L, Liu P, Xie Y, Choi J, Yu S, Lin Y, Dong J, Huang Q, Jin X, Lim M, Yang X. ACT001 reduces the expression of PD-L1 by inhibiting the phosphorylation of STAT3 in glioblastoma. Theranostics. 2020;10:5943–5956. doi: 10.7150/thno.41498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon J, Grinchuk OV, Tirado-Magallanes R, Ngian ZK, Tay EXY, Chuah YH, Lee BWL, Feng J, Crasta KC, Ong CT, Benoukraf T, Ong DST. E2F and STAT3 provide transcriptional synergy for histone variant H2AZ activation to sustain glioblastoma chromatin accessibility and tumorigenicity. Cell Death Differ. 2022;29:1379–1394. doi: 10.1038/s41418-021-00926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.