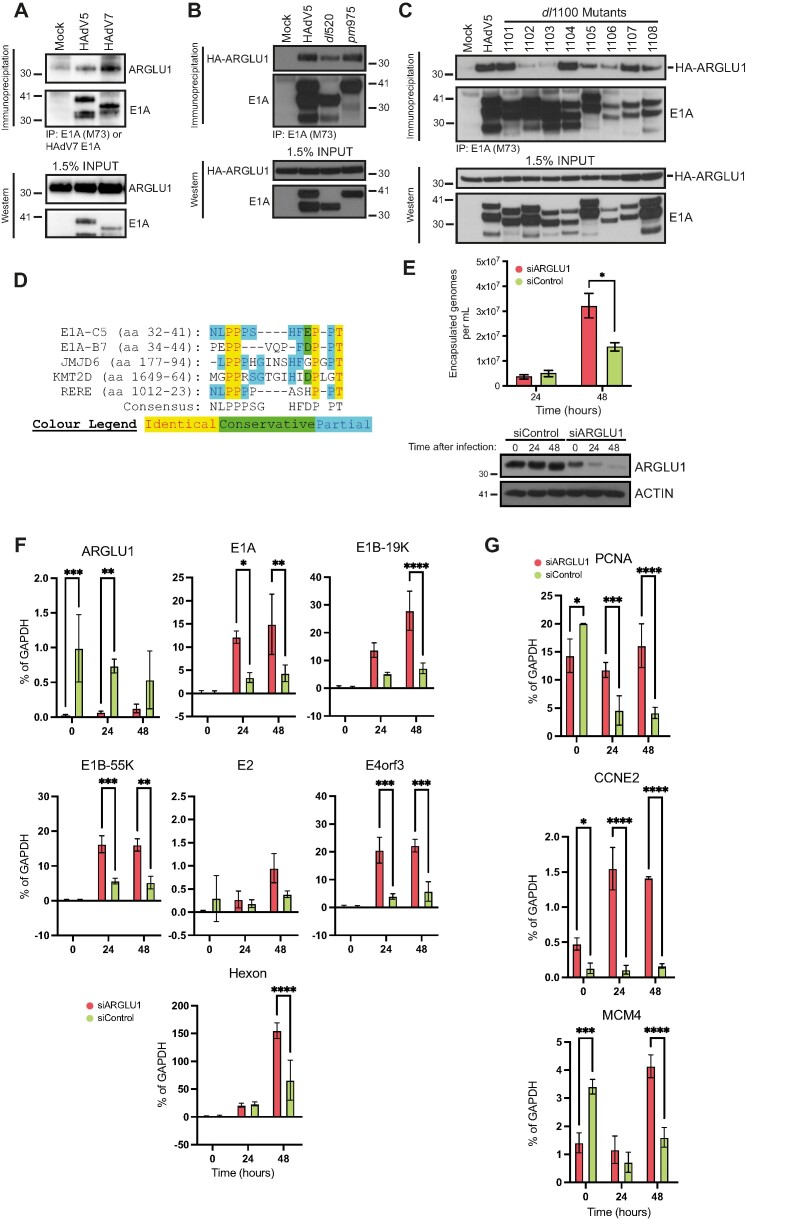
Figure 2.
ARGLU1 binds to E1A N-terminus and is a negative regulator of HAdV growth and gene expression. (A) HT cells were infected with HAdV-C5 dl309 or HAdV-B7 Gomen strain, immunoprecipitated 24 h after infection for E1A using M73 monoclonal antibody for HAdV-C5 and custom rabbit polyclonal antibody for HAdV-B7 E1A. Associated endogenous ARGLU1 was detected with a custom rabbit polyclonal anti-ARGLU1 antibody. (B) HT cells were transfected with HA-ARGLU1 and infected with the indicated dl309 HAdV-C5 or mutants pm975 expressing predominantly E1A289R or dl520 expressing predominantly E1A243R. Immunoprecipitations were performed 24 h after infection/transfection for E1A using M73 anti-E1A hybridoma and the associated HA-ARGLU1 was detected with anti-HA rat monoclonal antibody 3F10. (C) HT cells were infected with the indicated HAdV-C5 dl309-based deletion mutants and transfected with a plasmid expressing HA-ARGLU1, 24 h later cells were lysed and immunoprecipitated for E1A using the M73 monoclonal antibody. Associated HA-ARGLU1 was detected using rat anti-HA monoclonal antibody 3F10. (D) Multiple sequence alignment of the region in E1A that interacts with ARGLU1 and proteins with similar sequences. E1A-C5 refers to E1A from HAdV-C5 while E1A-B7 refers to HAdV-B7 E1A. (E) HT cells were transfected with siRNA targeting ARGLU1 mRNA (siARGLU1) or a negative control siRNA (siControl). Twenty-four hours later cells were infected with HAdV-C5, virus was allowed to replicate for 24 and 48 h prior to quantification. Viral particle counts were determined by quantifying encapsulated viral genomes, * P ≤ 0.05; n = 3. Bottom panel shows efficiency of ARGLU1 knockdown by siRNA at the 3 time points examined, time 0 is the time of virus infection and occurs 24 h after siRNA transfection. (F) Cells treated in the same way as in (B) above were harvested at the indicated time points for total RNA extraction using the TRIzol reagent. Time 0 represents uninfected cells. Total RNA was converted to cDNA using VILO reverse transcriptase master mix and used in qPCR to assay viral gene expression normalized to GAPDH mRNA levels; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001; n = 3. (G) Same as (D) except indicated cellular mRNAs were quantified; * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P ≤ 0.0001; n = 3.