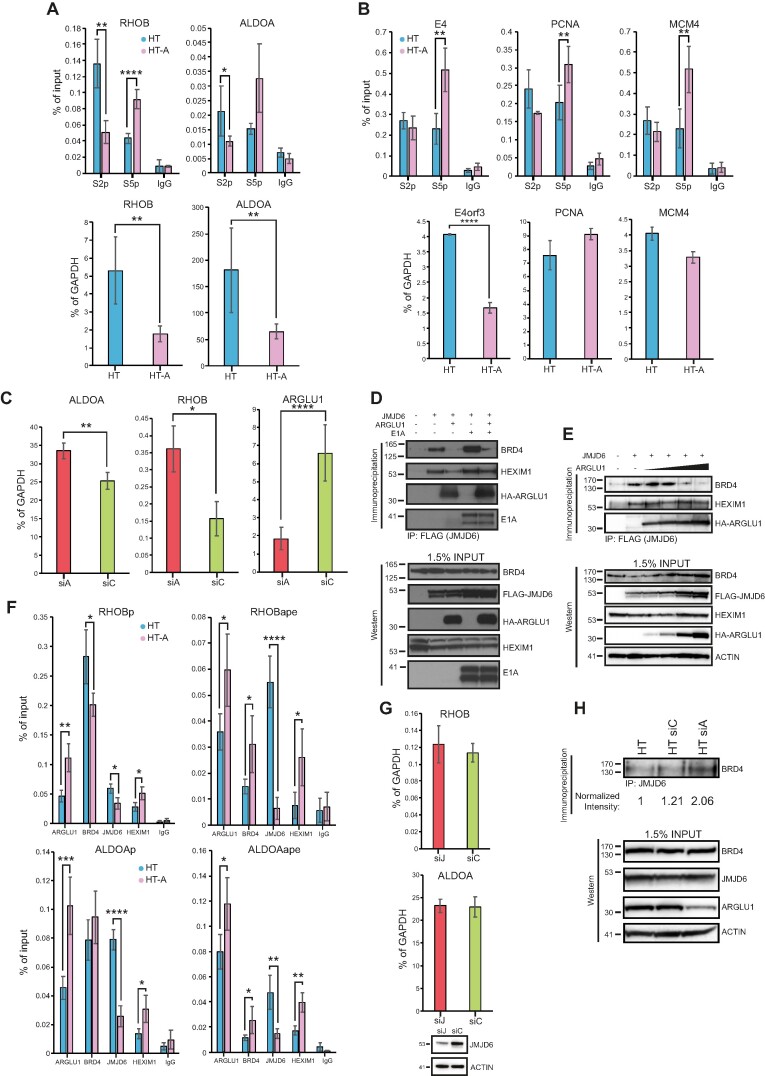
Figure 5.
ARGLU1 enhanced promoter proximal RNA polymerase II pausing. (A) Top panel; ChIPs of elongating (phosphorylated at the CTD at S2) or paused (phosphorylated at the CTD at S5) RNA polymerase II at the RHOB and ALDOA promoter-proximal region (+1 to +500 bp) in HT or HT-A cells; * P ≤ 0.05, ** P ≤ 0.01, **** P ≤ 0.0001; n = 4. IgG negative control consisted of rabbit anti-rat antibody. Bottom panel; expression levels of RHOB and ALDOA in HT or HT-A cells; ** P ≤ 0.01; n = 3. (B) Top panel, ChIPs of elongating (phosphorylated at the CTD at S2) or paused (phosphorylated at the CTD at S5) RNA polymerase II at the E4, PCNA and MCM4 promoter region in HT or HT-A cells. For the viral E4 gene the cells were infected for 24 h, for PCNA and MCM4 genes the cells were uninfected; ** P ≤ 0.01, n = 4. IgG negative control consisted of rabbit anti-rat antibody. Bottom panel; expression of E4orf3 in HAdV-C5 dl309-infected cells HT or HT-A cells, PCNA and MCM4 in uninfected HT and HT-A cells, cells were infected for assaying of viral E4orf3 expression, and uninfected for PCNA and MCM4; **** P ≤ 0.0001; n = 3. (C) HT cells were transfected with siRNA targeting ARGLU1 (siA) or a control siRNA (siC), mRNA levels of ALDOA, RHOB and ARGLU1 were assayed 48 h after siRNA transfection by extracting total RNA using TRIzol reagent, converting it to cDNA using VILO master mix reverse transcriptase and performing qPCR; * P ≤ 0.05, ** P ≤ 0.01, **** P ≤ 0.0001; n = 3. (D) HT cells were transfected with the indicated plasmids to express FLAG-tagged JMJD6, HA-ARGLU1, and HAdV-C5 E1A. Cells were then lysed and immunoprecipitated for FLAG using M2 FLAG resin to precipitate JMJD6 and associated proteins. Western blots were then performed for BRD4, HEXIM1, HA-ARGLU1, E1A and FLAG-JMJD6 on resolved immunoprecipitations and inputs, HA-ARGLU1 and FLAG-JMJD6 were detected using anti-HA rat monoclonal antibody 3F10, FLAG-JMJD6 is double-tagged with HA and FLAG epitopes, the remaining proteins were detected using their specific antibodies described in the materials and methods. (E) HT cells were transfected with a plasmid to express FLAG-tagged JMJD6 and increasing concentrations (0, 0.5, 1, 2 and 5 μg) of a plasmid expressing HA-ARGLU1. Twenty-four hours later, cells were lysed and immunoprecipitated for FLAG using M2 FLAG resin to precipitate JMJD6 and associated proteins. Western blots were then performed for ARGLU1, BRD4, HEXIM1, HA-ARGLU1, and FLAG-JMJD6 on resolved immunoprecipitations and inputs (as in D). ACTIN was used as a loading control. (F) HT or HT-A cells were subjected to ChIP analysis with the indicated antibodies to assess occupancy of ARGLU1, BRD4, JMJD6 and HEXIM1 at the promoters (P; –1000 to +1) and anti-pause enhancers (ape) of RHOB and ALDOA genes. Non-specific rabbit anti-rat immunoglobulin (IgG) was used as a negative control; * P ≤ 0.05, ** P ≤ 0.01, **** P ≤ 0.0001; n = 3. (G) HT-A cells were transfected with an siRNA targeting JMJD6 (siJ) or a negative control siRNA (siC) for 48 h, at which time total RNA was extracted using TRIzol method, converted to cDNA using VILO master mix, and analyzed for the levels of RHOB and ALDOA mRNA by qPCR. GAPDH was used as a normalization control and the data is represented as percentage of GAPDH. Bottom blot shows knockdown of JMJD6 48 h after siRNA transfection. (H) HT cells were left untreated or transfected with a control siRNA (siC) or ARGLU1 siRNA (siA) for 48 h. Cells were then lysed and immunoprecipitated for JMJD6. Immunoprecipitates were resolved on SDS polyacrylamide gel and probed for BRD4. Density of BRD4 bands was quantified using AzureSpot software and normalized to total BRD4 and ACTIN loading controls and is represented as fold increase over untreated HT cells. Inputs are shown for BRD4, JMJD6, ARGLU1 and ACTIN.